Template for Nucleic Acids Research (NAR) journal
Originally from http://www.oxfordjournals.org/our_journals/nar/for_authors/msprep_submission.html


Get in touch
Have you checked our knowledge base ?
Message sent! Our team will review it and reply by email.
Email:
An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Copyright and License information
This is an Open Access article distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted reuse, distribution, and reproduction in any medium, provided the original work is properly cited.
Graphical Abstract.
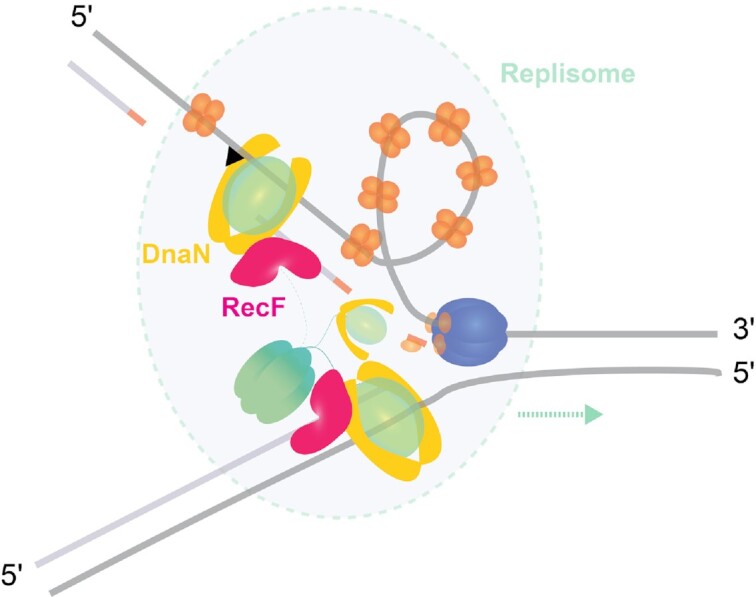
RecF interacts with DnaN.
KEGG: Kyoto Encyclopedia of Genes and Genomes
Affiliation.
- 1 Institute for Chemical Research, Kyoto University, Uji, Kyoto 611-0011, Japan.
- PMID: 9847135
- PMCID: PMC148090
- DOI: 10.1093/nar/27.1.29
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a knowledge base for systematic analysis of gene functions in terms of the networks of genes and molecules. The major component of KEGG is the PATHWAY database that consists of graphical diagrams of biochemical pathways including most of the known metabolic pathways and some of the known regulatory pathways. The pathway information is also represented by the ortholog group tables summarizing orthologous and paralogous gene groups among different organisms. KEGG maintains the GENES database for the gene catalogs of all organisms with complete genomes and selected organisms with partial genomes, which are continuously re-annotated, as well as the LIGAND database for chemical compounds and enzymes. Each gene catalog is associated with the graphical genome map for chromosomal locations that is represented by Java applet. In addition to the data collection efforts, KEGG develops and provides various computational tools, such as for reconstructing biochemical pathways from the complete genome sequence and for predicting gene regulatory networks from the gene expression profiles. The KEGG databases are daily updated and made freely available (http://www.genome.ad.jp/kegg/).
Publication types
- Research Support, Non-U.S. Gov't
- Computational Biology
- Databases, Factual*
- Gene Expression
- Sequence Homology
- Search Menu
- Sign in through your institution
- Chemical Biology and Nucleic Acid Chemistry
- Computational Biology
- Critical Reviews and Perspectives
- Data Resources and Analyses
- Gene Regulation, Chromatin and Epigenetics
- Genome Integrity, Repair and Replication
- Molecular and Structural Biology
- Nucleic Acid Enzymes
- Nucleic Acid Therapeutics
- RNA and RNA-protein complexes
- Synthetic Biology and Bioengineering
- Advance Articles
- Breakthrough Articles
- Molecular Biology Database Collection
- Special Collections
- Scope and Criteria for Consideration
- Author Guidelines
- Data Deposition Policy
- Database Issue Guidelines
- Web Server Issue Guidelines
- Submission Site
- Open Access Charges
- About Nucleic Acids Research
- Editors & Editorial Board
- Information of Referees
- Self-Archiving Policy
- Dispatch Dates
- Advertising and Corporate Services
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic
Article Contents
Introduction, materials and methods, data availability, acknowledgements.
- < Previous
RNAcanvas: interactive drawing and exploration of nucleic acid structures
- Article contents
- Figures & tables
- Supplementary Data
Philip Z Johnson, Anne E Simon, RNAcanvas: interactive drawing and exploration of nucleic acid structures, Nucleic Acids Research , Volume 51, Issue W1, 5 July 2023, Pages W501–W508, https://doi.org/10.1093/nar/gkad302
- Permissions Icon Permissions
Two-dimensional drawing of nucleic acid structures, particularly RNA structures, is fundamental to the communication of nucleic acids research. However, manually drawing structures is laborious and infeasible for structures thousands of nucleotides long. RNAcanvas automatically arranges residues into strictly shaped stems and loops while providing robust interactive editing features, including click-and-drag layout adjustment. Drawn elements are highly customizable in a point-and-click manner, including colours, fonts, size and shading, flexible numbering, and outlining of bases. Tertiary interactions can be drawn as draggable, curved lines. Leontis-Westhof notation for depicting non-canonical base-pairs is fully supported, as well as text labels for structural features (e.g. hairpins). RNAcanvas also has many unique features and performance optimizations for large structures that cannot be correctly predicted and require manual refinement based on the researcher's own analyses and expertise. To this end, RNAcanvas has point-and-click structure editing with real-time highlighting of complementary sequences and motif search functionality, novel features that greatly aid in the identification of putative long-range tertiary interactions, de novo analysis of local structures, and phylogenetic comparisons. For ease in producing publication quality figures, drawings can be exported in both SVG and PowerPoint formats. URL: https://rnacanvas.app .

Exploring the 2,692 nt citrus yellow vein-associated virus genome structure in RNAcanvas. Currently selected sequence highlighted yellow. RNAcanvas has highlighted two putative tertiary pairings purple (hovered by cursor) and pink.
In nucleic acids research, particularly RNA research, structures are commonly drawn in 2-dimensional (2D) format ( 1–3 ). Conventions such as the Leontis-Westhof notation ( 4 ) are also important for denoting various types of canonical and non-canonical base-pairs and interactions in RNA structures. Since manually creating 2D structure drawings is laborious for large structures ( 5–7 ), a variety of software programs have been developed to facilitate this process.
2D nucleic acid structure drawing programs can be roughly separated into two categories: (i) programs that offer flexible, force-based adjustment of drawing layouts and (ii) programs that generally apply stricter layout conventions to drawings. Programs in the first category, including forna ( 8 ) and RiboSketch ( 9 ), allow click-and-drag editing of drawings and use force-based methods ( 8 ) to help maintain the arrangement of residues into stems and loops in response to dragging. The interactivity features in this class of programs make editing drawings intuitive, and their use of force-based methods allow for great flexibility in how a drawing is arranged. These programs, however, require increased effort to (for example): (i) precisely align stems emanating from a junction or that are stacked; (ii) flatten or evenly round regions of unpaired residues and (iii) align stems with the flat base of a drawing.
Programs in the second category, including VARNA ( 10 ), XRNA ( http://rna.ucsc.edu/rnacenter/xrna/xrna_faq.html ), PseudoViewer ( 11 ), RnaViz ( 12 ), jViz.RNA ( 13 ), RNAView ( 14 ), RNApuzzler ( 15 ), R2R ( 16 ), R2DT ( 17 ) and RNArtist ( https://github.com/fjossinet/RNArtist ), generally produce drawings in a more regimented, publication-friendly manner that can more efficiently communicate secondary structure and facilitate phylogenetic comparisons ( 2 , 15–18 ). Importantly, programs in this category have greater functionality for precisely arranging stems and loops in a strict manner (e.g. perfectly round or flattened loops), making them preferred for the drawing of large structures, which benefit from stricter layout organization ( 15 , 17 ). These programs also introduce additional methods for specifying the aspects of a drawing, such as program-specific input file formats (e.g. R2R, RNArtist) and layout templates (e.g. R2DT), and also tend to offer greater customization of individual drawing elements including bases and bond notations (e.g. Leontis-Westhof). On the other hand, features for interactivity, such as click-and-drag editing of residue layout and the shapes of tertiary interaction lines, are often lacking or limited in functionality. This category of programs may also lack point-and-click editing of base-pairs and other drawing elements, thus requiring input files or application forms for edits, where bases and other elements are specified by numeric position making editing drawings cumbersome and less intuitive.
Nucleic acid structure drawings are rarely static but change as substructures are confirmed using biochemical methods or through phylogenetic comparisons, and thus drawings should also be easily editable to reflect a researcher's current knowledge. Towards this goal, RNAcanvas was designed to combine some of the best features of both types of structure drawing programs, merging the interactivity functions of the first category (e.g. click-and-drag layout adjustments) with the orderly layouts (e.g. residues maintained in strictly shaped stems and loops) and detailed drawing customizations of the second category. Furthermore, RNAcanvas provides extensive point-and-click editing features for individual components of the drawing, such as base letters, lines of bonds, and base outlines and numberings. RNAcanvas is also one of the few programs to fully support Leontis-Westhof notation for depicting canonical and non-canonical RNA base-pairs and interactions. RNAcanvas has been optimized to function smoothly with large structures, and contains unique features such as real-time highlighting of complementary sequences, motif search, and export of drawings in both scalable vector graphics (SVG) and PowerPoint (PPTX) formats, which greatly facilitates the creation of figures for scientific presentations. Exported SVG and PPTX drawings are also formatted such that individual objects (e.g. base letters, bond lines) can be altered in vector graphics editors such as Adobe Illustrator or PowerPoint. RNAcanvas was formerly named RNA2Drawer, which was initially available only in a download format when first published ( 19 ).
Versions of software tools used for RNAcanvas are currently the most recent (2023), however continuous enhancements to the program to provide additional functionality and updating of tools will continue.
Programming languages and general tools
RNAcanvas is a single-page web application created with HTML/CSS/JavaScript. Babel v7.16.10 is used to compile JavaScript code to the ECMAScript 2009 (ES5) standard. The application is built using the webpack module bundler v4.46.0. TypeScript v3.9.10 is used to add type annotations over JavaScript code. All JavaScript application code to be received by the user is written with TypeScript type annotations. Unit testing is performed on the Node.js runtime environment v16.18.1 and using the Jest framework v24.9.0. The uuid library v8.3.2 is used to generate universally unique identifiers (UUIDs), which are assigned to a variety of components (e.g. text elements of bases) to simplify retrieval in the application code.
Interactive drawing
Within the application, the drawing itself is a scalable vector graphics (SVG) document, which is a type of element that web browsers can render. Web browsers provide built-in methods for detecting user interaction with SVG documents (e.g. mouse clicks, hovering and dehovering of elements within the SVG document). This allows user interaction with the drawing to be programmed in an event-driven manner. The SVG specification is mature and extensive, allowing additional customization of drawings. The SVG.js library v3.1.1 is also used to simplify code for manipulating the drawing and managing user interactions.
Peripheral UI elements
Peripheral user interface (UI) elements, such as the top menu, bottom information bar, and right-side forms are built using the React framework v16.14.0. The react-color library v2.19.3 is used for the colour picker UI element. The react-select library v3.2.0 is used for the UI element for picking fonts. The React Testing Library v9.5.0 and the enzyme library v3.11.0 are used for unit testing of peripheral UI elements.
Exporting drawings
The SVG.js library v3.1.1 is used to produce SVG files of drawings. (SVG files contain the textual representation of an SVG document, such as the drawing of the application.) To produce PowerPoint files of drawings, the PptxGenJS library v3.9.0 is used. Rather than convert all the SVG elements of a drawing to their nearest PowerPoint object counterpart, many drawing elements (e.g. the curved lines of tertiary bonds) are included in PPTX files as individual SVG image elements. Because of this, exported PPTX files require PowerPoint version 2016 or later. SVG image elements in exported PPTX files can still be manipulated as though they were PowerPoint objects and can often be converted to corresponding PowerPoint objects using the PowerPoint ‘Convert to Shape’ feature.
RNAcanvas offers extensive options to customize drawings of nucleic acid secondary structures and tertiary interactions based on graphical user interaction, with drawings edited using ‘tools’ and forms. Tools control how the user interacts with the drawing itself. For example, different tools allow the user to drag elements of the drawing, select element(s) for editing, or pair and unpair bases. Forms pop-up on the right-side of the application and allow properties of selected elements (or the drawing as a whole) to be directly edited through text and numeric inputs and various picker components and toggles (e.g. letters and fonts of bases, colours of bonds, base numbering). All structure drawing figures in this report were produced entirely within the RNAcanvas web app (unless stated otherwise).
Layout flexibility
Using the Dragging Tool , the layout of a drawing can be adjusted by dragging with the mouse (Figure 1 ). Stems can be dragged around loops and the outermost loop can be dragged to rotate the drawing with the arrangement of bases into stems and loops strictly maintained. Using the Flattening Tool , individual loops can be flattened (or reverted to unflattened). When flattened, the outermost loop is entirely flat (Figure 1B , a - c ), and inner loops with more than one child stem assume a triangular shape (Figure 1B , d – g ). Flattening inner loops with multiple child stems often helps to condense a drawing and highlight conserved branching patterns in structures. Flattening an inner loop with only one child stem (Figure 1B , h and i ) aligns the child stem with the parent stem, effectively ‘straightening’ the two stems. Using the Flipping Tool , individual stems can be flipped across their parent loops (Figure 1B , j – m ). RNAcanvas also has a Layout form in which the rotation of the drawing can be precisely specified. The Layout form also has a field for specifying the termini gap of the drawing, which is the distance between the first and last bases when the outermost loop of a drawing is round (Figure 1B , label n and dashed line). The termini gap can be set to zero (for a fully circular outermost loop) or to larger values (e.g. for a semicircle outermost loop).

Various layout options for drawings in RNAcanvas. ( A ) The same input structure as in (B, left) before any layout adjustments save for being rotated 90°. ( B ) Labels a-n and dashed line (lower right) added in Adobe Illustrator. (a) Flat outermost loop. (b) Rotated flat outermost loop. (c) Round outermost loop. (d, e) Round inner loops. (f, g) Flattened inner loops with multiple child stems. (h) Flattened inner loop with only one child stem that has been condensed. (i) Flattened inner loop with only one child stem that has been stretched. (j-m) Flipped stems. (n) Termini gap of the round outermost loop (indicated by dashed line).
Customization of drawing elements
The following customizations can be accomplished using the Editing Tool :
Base customizations . Fonts, font sizes and base colours can be personalised (Figure 2A ). Bases can be outlined, and outlines can be adjusted for size, line thickness and line and fill colours and transparencies. Chemical probing data such as SHAPE data ( 20 ) can be easily incorporated into the structure through base and/or outline colouring. A ‘dots’ style drawing, in which only the colour-coded outlines of bases are shown, is an effective way to communicate chemical probing data (Figure 2B ) and dots can be easily suppressed to reveal the underlying bases. RNAcanvas has a dedicated form to help with styling bases according to a set of data. This form allows the user to progressively select and style bases within different data ranges (e.g. low, moderate, and high SHAPE reactivities). Base letters can be directly edited, and bases can subsequently be inserted, appended, and removed. A substructure can be specified when inserting/appending a subsequence of bases. The numbering of bases can also be specified via the offset, increment and anchor properties in the Numbering form , with text and lines customizable in terms of fonts, colour, dimensions, etc. (Figure 2A ). Individual bases can also be manually numbered regardless of what numbers are assigned to other bases in the drawing.
Bond customizations . In a drawing, consecutive bases are linked by a primary bond. Most primary bonds are not visible (by default) when a structure is initially drawn. Secondary bonds join base-pairs in the secondary structure of a drawing and affect the layout of the drawing. Tertiary bonds communicate base-pairs/interactions that do not affect the layout of the drawing. All bonds have a line that connects the two bases in the bond. Primary and secondary bonds have straight lines. Tertiary bonds have curved lines that can be dragged with the mouse to adjust their shape. The lines of bonds can be customized in terms of colour and thickness, as well as base padding, which is the distance between the end of a bond line and the base connected by the bond (Figure 2C ). Bond lines can be dashed and their ends rounded.

Various customizations of drawing elements. ( A ) Fonts, font sizes and colours of bases text are all customizable. Bases can be outlined with circles and numbered. Base outlines and numberings are also customizable in terms of colours, dimensions, fonts, etc. This drawing has a base numbering offset of –121 nt. ( B ) Drawing presented in ‘dots’ style in which only colour-coded outlines of bases are shown. Dots style is an effective way to communicate chemical probing data, such as the example SHAPE data shown in this drawing. Dots colour legend added in Adobe Illustrator. ( C ) Various customizations of bond lines and ‘strung’ elements. Entire Leontis-Westhof notation for depicting canonical and non-canonical base-pairs and interactions is supported. ( D ) Text labels and base markers. Hairpin H1, stem–loop SL2, linker region L3, the 5′end and the 3′OH were all labelled within the program. Bases also can have various markers (i.e. coloured triangles, squares, circles) associated with them. These text labels and base markers are ‘strung’ elements of nearby primary and secondary bonds that have been dragged to their current positions. Due to being strung elements, these text labels and base markers maintain both their positions and orientations relative to their parent bonds when the layout of the drawing is adjusted. ( E ) Shading sequences of bases. Tertiary pairing connected by a single tertiary bond is shown (orange). ( F ) Drawing with portions outlined. ( G ) Drawing with portions in the ‘stick-and-ball’ style.
Elements such as rectangles, circles, triangles, and text can be ‘strung’ onto (associated with) any bond (Figure 2C ). This attachment of elements to specific bonds allows for the full Leontis-Westhof notation ( 4 ) for canonical and non-canonical base-pairs and interactions to be depicted on secondary and tertiary bonds. As with other drawing elements, strung elements can be customized in terms of colour, transparencies, dimensions, line dashing, fonts, etc. Strung elements can be dragged with the mouse and displaced from their parent bond, which adds versatility to their usage. For example, when dragged next to nearby bases, strung elements can mark the location with additional information (Figure 2D ) or can be used to shade hairpins. Text strung elements can be used to name structural features as well as the 5′ and 3′ termini (Figure 2D ). All strung elements maintain their position and orientation relative to their parent bond when the layout of the drawing is adjusted.
Shading a series of bases and outlining structures . Highlighting a selection of bases can be accomplished by: (i) setting primary bond base paddings to zero; (ii) making primary bonds sufficiently thick to encompass the font size of the bases; (iii) setting the line end caps of the primary bonds to round and (iv) sending the shading below the bases (Figure 2E ). Tertiary pairings can also be denoted by shading the two interacting sequences and connecting one base of each sequence with a single tertiary bond (Figure 2E , in orange). It is also possible to outline structures in a drawing by shading bases using this technique and then sending base letters below the shading (Figure 2F ).
‘Stick-and-ball’ drawings . ‘Stick-and-ball’ drawings (Figure 2G ) can be created by making base letters invisible (e.g. by assigning them the background colour), increasing the base padding of secondary bonds to shorten them, and making secondary bonds sufficiently thick to overlap with each other (thus forming the ‘stick’ portion). The ‘ball’ portion is composed of circle strung elements that are dragged over the loops in the structure and sized to match the sizes of the loops.
Structure editing and large structure exploration
Structures thousands of nucleotides long, such as the structures of positive-strand RNA virus genomes, are increasingly studied as a whole ( 5 , 6 , 21 ). Such large structures cannot be predicted directly ( 5 ) and require manual refinement by RNA researchers based on phylogenetic comparisons, experimental results, and the researcher's own expertise. RNAcanvas has optimized performance for editing large structures, e.g. when dragging to adjust the layout.
The Pairing Tool is used to pair and unpair bases (i.e. add and remove secondary and tertiary bonds) in a point-and-click manner. The Pairing Tool also has the option of highlighting sequences that are complementary to the selected sequence (Figure 3 , selected sequence highlighted in yellow and complementary sequences highlighted in pink). The number of possible pairing partners within the structure can be designated, and options are available to exclude G-U/G-T pairs and permit partial mismatches between pairing partners. The Pairing Tool aids in refinement of output by structure prediction programs such as Mfold ( 22 ) and RNAfold ( 23 ) and helps to identify possible long-range base-pairings that cannot currently be predicted computationally ( 5 ). RNAcanvas also has a form for finding input motifs ( Find Motifs form ) (Figure 3 ). Both complement highlighting (using the Pairing Tool) and the Find Motifs form support partial mismatching and the use of IUPAC single letter codes ( 24 ). Complement highlighting further supports a percentage cap on the G-U/G-T base-pair composition of highlighted complements. The Find Motifs form can also treat an input motif to search for as a regular expression (as defined in Computer Science).

Screenshot showing highlighting of complementary sequences with the Pairing Tool and the Find Motifs form. The 5′CAACC sequence highlighted in yellow is currently selected. Complementary sequences 5′GGUUG found by the program are highlighted in pink. (Right) Find Motifs form is open. Currently the options to match Us and Ts and to use IUPAC single letter codes are toggled. The motif 5′CYRCVA (containing the IUPAC single letter codes Y, R and V) was searched for and six matching motifs were found. The second matching motif from the bottom is being hovered with the mouse cursor (highlighted in grey). Clicking on a matching motif will centre the matching motif on the screen and flash its base letters.
Input and output formats
RNAcanvas accepts primary sequences (unstructured) as well as structures in dot-bracket notation (also called Vienna format) and in Connectivity Table (CT) files. Input structures may contain pseudoknots, which are drawn using tertiary bonds. RNAcanvas outputs drawings in SVG and PowerPoint formats. SVG files can be opened in vector graphics editors such as Adobe Illustrator and Inkscape. Drawings are exported such that all drawing elements (e.g. bases text, bond lines) are exported as individual SVG or PowerPoint objects. For instance, base letters are exported as SVG text elements or PowerPoint text boxes. This allows for further manipulation of exported drawings in vector graphics editors and PowerPoint. A scaling factor may also be specified when exporting a drawing. All elements in an exported drawing are individually scaled according to the scaling factor. Scaling elements individually on export eases the manipulation of exported drawings compared to when exported drawings are scaled as a whole after export using a separate application such as Adobe Illustrator. Additionally, RNAcanvas can save drawings in a file format unique to RNAcanvas with ‘.rnacanvas’ extension. Downloaded drawing files contain a complete representation of a drawing, allowing drawings to be reopened at later times exactly as they were when saved.
RNAcanvas has many enhancements over the free desktop-based RNA2Drawer predecessor ( 19 ). Being web-based, RNAcanvas is easier for users to access and is still available for use at no cost. The RNAcanvas web app also uses the SVG web standard to present drawings, which is expansive and permits wide customization of drawings. Other improvements in the web app include unlimited undo/redo functionality, greater layout flexibility (e.g. the ability to flatten and circularize loops individually), the availability of strung elements on bonds, support for Leontis-Westhof notation, ability to add text labels and base markers, enhanced flexibility in numbering bases, and the Find Motifs form.
Although RNAcanvas offers flexible options to customize the layout of a drawing (Figure 1 ), the program does not currently support certain layout features. For instance, RNAcanvas cannot currently incorporate pseudoknots into the layout of a drawing, unlike some programs such as PseudoViewer (Table 1 ) ( 11 ). Certain types of structures such as frameshifting elements ( 25 ) are often drawn such that pseudoknot(s) are conveyed via the layout of the drawing. The initial layouts of drawings made by RNAcanvas (especially for large structures) can also contain overlapping regions that need to be manually untangled by the user. While drawings produced by many programs often contain overlapping regions ( 17 ), some programs such as RNApuzzler/RNAturtle have made algorithmic advancements in the initial layout of an RNA drawing to avoid overlaps ( 15 ). R2DT has also demonstrated the effectiveness of template-based drawing for a wide array of small and large structures ( 17 ). Programs such as PseudoViewer, RNApuzzler and R2DT, however, do not provide the expansive features of RNAcanvas for editing drawings, such as click-and-drag layout adjustment and point-and-click editing of the structure and individual drawing elements, which allow structure drawings to be moulded beyond their initial forms. RNAcanvas is also unique in its focus on the extended exploration of structures and the continued editing of drawings that occurs as the researcher's knowledge of a structure grows.
Comparison of programs for 2D drawing of nucleic acid structures. Solid dots indicate that a feature is supported by a program, though the robustness of feature implementations may vary between different programs. VARNA and jViz.RNA have hollow dots for web-based use since they require a Java plugin for use in a web browser. Point-and-click structure editing refers to the ability to add and remove base-pairs and interactions in the secondary and tertiary structures of a drawing graphically using the mouse. Motif finding functionality refers to features similar to the Find Motifs form in RNAcanvas. Point-and-click element editing is the ability to select individual or groups of elements to edit by clicking on them and/or dragging a selecting box over them, rather than being required to select elements by less direct means (e.g. by inputting numeric positions). Text labels for structural features refer to the ability to place pieces of text next to structural features such as hairpins and linker regions that name them. Flexible base numbering is the ability to assign any number to any base regardless of what numbers are assigned to other bases. This is useful when a drawing contains two discontinuous portions of a larger sequence and the numbering for one portion must be offset from the other. Granular output scaling refers to the ability to scale drawings at the level of individual elements when exported, as opposed to scaling an exported drawing as a whole after export. Drawing using input files and automated drawing are grouped together since the use of input files can lend itself to automated drawing via the automated creation of input files. Automated drawing also includes the ability to generate large numbers of structure drawings in an automated way in general, such as by allowing other software written by the user to interface with the structure drawing software, such as with VARNA
RNAcanvas is able to handle large (at least 3000 nt) structures with only modest lag times, making it significantly faster than other drawing platforms with similar click-and-drag layout adjustment. The primary performance bottleneck of RNAcanvas is the re-rendering of base letters when the layout of a drawing is changed. Note that among Chrome, Firefox and Safari, the Chrome web browser currently has the best performance when handling large RNAcanvas structures. Many biological transcripts, such as positive-strand RNA virus genomes and long noncoding RNAs, have lengths over 3 kb and thus future performance improvements will need to include new strategies for minimizing lag times when dragging portions of very large structures. Ultimately, RNAcanvas aspires to allow entire structures of any size biological transcript to be effectively drawn and edited on personal devices.
RNAcanvas web app: https://rnacanvas.app . RNAcanvas GitHub repository: https://github.com/pzhaojohnson/rnacanvas and https://doi.org/10.6084/m9.figshare.22134029.v1 .
We would like to thank Dr Bruce Shapiro and Dr Wojciech Kasprzak for their invaluable help in developing RNA2Drawer, the predecessor to RNAcanvas, and all of the advice that they have provided over the years that has propelled this project forward.
National Science Foundation [MCB-1818229]; United States Department of Agriculture [308291-00001 to A.E.S.]; P.Z.J. was partially supported by National Science Foundation Graduate Fellowship Award [DGE-1840340]. Funding for open access charge: USDA [308291-00001].
Conflict of interest statement . None declared.
Ponty Y. , Leclerc F. Drawing and editing the secondary structure(s) of RNA . Methods Mol. Biol. 2015 ; 1269 : 63 – 100 .
Google Scholar
Shabash B. , Wiese K.C. RNA visualization: relevance and the current state-of-the-art focusing on pseudoknots . IEEE/ACM Trans. Comput. Biol. Bioinform. 2017 ; 14 : 696 – 712 .
Vicens Q. , Kieft J.S. Thoughts on how to think (and talk) about RNA structure . Proc. Natl. Acad. Sci. U.S.A. 2022 ; 119 : e2112677119 .
Leontis N.B. , Westhof E. Geometric nomenclature and classification of RNA base pairs . RNA . 2001 ; 7 : 499 – 512 .
Lu Z. , Chang H.Y. The RNA base-pairing problem and base-pairing solutions . Cold Spring Harb. Perspect. Biol. 2018 ; 10 : a034926 .
Liu J. , Carino E. , Bera S. , Gao F. , May J.P. , Simon A.E. Structural analysis and whole genome mapping of a new type of plant virus subviral RNA: umbravirus-like associated RNAs . Viruses . 2021 ; 13 : 646 .
Chkuaseli T. , White K.A. Activation of viral transcription by stepwise largescale folding of an RNA virus genome . Nucleic Acids Res. 2020 ; 48 : 9285 – 9300 .
Kerpedjiev P. , Hammer S. , Hofacker I.L. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams . Bioinformatics . 2015 ; 31 : 3377 – 3379 .
Lu J.S. , Bindewald E. , Kasprzak W.K. , Shapiro B.A. RiboSketch: versatile visualization of multi-stranded RNA and DNA secondary structure . Bioinformatics . 2018 ; 34 : 4297 – 4299 .
Darty K. , Denise A. , Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure . Bioinformatics . 2009 ; 25 : 1974 – 1975 .
Byun Y. , Han K. PseudoViewer3: generating planar drawings of large-scale RNA structures with pseudoknots . Bioinformatics . 2009 ; 25 : 1435 – 1437 .
De Rijk P. , Wuyts J. , De Wachter R. RnaViz 2: an improved representation of RNA secondary structure . Bioinformatics . 2003 ; 19 : 299 – 300 .
Shabash B. , Wiese K.C. jViz.RNA 4.0-Visualizing pseudoknots and RNA editing employing compressed tree graphs . PLoS One . 2019 ; 14 : e0210281 .
Yang H. , Jossinet F. , Leontis N. , Chen L. , Westbrook J. , Berman H. , Westhof E. Tools for the automatic identification and classification of RNA base pairs . Nucleic Acids Res. 2003 ; 31 : 3450 – 3460 .
Wiegreffe D. , Alexander D. , Stadler P.F. , Zeckzer D RNApuzzler: efficient outerplanar drawing of RNA-secondary structures . Bioinformatics . 2019 ; 35 : 1342 – 1349 .
Weinberg Z. , Breaker R.R. R2R–software to speed the depiction of aesthetic consensus RNA secondary structures . BMC Bioinf. 2011 ; 12 : 3 .
Sweeney B.A. , Hoksza D. , Nawrocki E.P. , Ribas C.E. , Madeira F. , Cannone J.J. , Gutell R. , Maddala A. , Meade C.D. , Williams L.D. et al. . R2DT is a framework for predicting and visualising RNA secondary structure using templates . Nat. Commun. 2021 ; 12 : 3494 .
Muller G. , Gaspin C. , Etienne A. , Westhof E. Automatic display of RNA secondary structures . Comput. Appl. Biosci. 1993 ; 9 : 551 – 561 .
Johnson P.Z. , Kasprzak W.K. , Shapiro B.A. , Simon A.E. RNA2Drawer: geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences . RNA Biol . 2019 ; 16 : 1667 – 1671 .
Wilkinson K.A. , Merino E.J. , Weeks K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution . Nat. Protoc. 2006 ; 1 : 1610 – 1616 .
Ziv O. , Price J. , Shalamova L. , Kamenova T. , Goodfellow I. , Weber F. , Miska E.A. The short- and long-range RNA-RNA interactome of SARS-CoV-2 . Mol. Cell . 2020 ; 80 : 1067 – 1077 .
Zuker M. Mfold web server for nucleic acid folding and hybridization prediction . Nucleic Acids Res. 2003 ; 31 : 3406 – 3415 .
Lorenz R. , Bernhart S.H. , Höner Zu Siederdissen C. , Tafer H. , Flamm C. , Stadler P.F. , Hofacker I.L. ViennaRNA Package 2.0 . Algorithms Mol. Biol. 2011 ; 6 : 26 .
Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984 . Nucleic Acids Res. 1985 ; 13 : 3021 – 3030 .
Firth A.E. , Brierley I. Non-canonical translation in RNA viruses . J. Gen. Virol. 2012 ; 93 : 1385 – 1409 .
Email alerts
Citing articles via.
- Editorial Board
- X (formerly Twitter)
Affiliations
- Online ISSN 1362-4962
- Print ISSN 0305-1048
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 23 October 2024
SpliceTransformer predicts tissue-specific splicing linked to human diseases
- Ningyuan You ORCID: orcid.org/0009-0000-3630-1604 1 ,
- Chang Liu 1 ,
- Yuxin Gu ORCID: orcid.org/0009-0009-5518-3493 2 ,
- Rong Wang 3 ,
- Hanying Jia 1 ,
- Tianyun Zhang 1 ,
- Song Jiang ORCID: orcid.org/0000-0002-7656-9434 4 ,
- Jinsong Shi 4 ,
- Ming Chen ORCID: orcid.org/0000-0002-9677-1699 5 ,
- Min-Xin Guan ORCID: orcid.org/0000-0001-5067-6736 2 ,
- Siqi Sun ORCID: orcid.org/0000-0001-7240-8724 6 ,
- Shanshan Pei 1 , 7 ,
- Zhihong Liu ORCID: orcid.org/0000-0001-6093-0726 4 &
- Ning Shen ORCID: orcid.org/0000-0003-4709-3374 1
Nature Communications volume 15 , Article number: 9129 ( 2024 ) Cite this article
1 Altmetric
Metrics details
- Computational models
- Genetic counselling
- Machine learning
- RNA splicing
- Sequence annotation
We present SpliceTransformer (SpTransformer), a deep-learning framework that predicts tissue-specific RNA splicing alterations linked to human diseases based on genomic sequence. SpTransformer outperforms all previous methods on splicing prediction. Application to approximately 1.3 million genetic variants in the ClinVar database reveals that splicing alterations account for 60% of intronic and synonymous pathogenic mutations, and occur at different frequencies across tissue types. Importantly, tissue-specific splicing alterations match their clinical manifestations independent of gene expression variation. We validate the enrichment in three brain disease datasets involving over 164,000 individuals. Additionally, we identify single nucleotide variations that cause brain-specific splicing alterations, and find disease-associated genes harboring these single nucleotide variations with distinct expression patterns involved in diverse biological processes. Finally, SpTransformer analysis of whole exon sequencing data from blood samples of patients with diabetic nephropathy predicts kidney-specific RNA splicing alterations with 83% accuracy, demonstrating the potential to infer disease-causing tissue-specific splicing events. SpTransformer provides a powerful tool to guide biological and clinical interpretations of human diseases.
Similar content being viewed by others
A deep learning approach to identify gene targets of a therapeutic for human splicing disorders
Mechanism and modeling of human disease-associated near-exon intronic variants that perturb RNA splicing
Aberrant splicing prediction across human tissues
Introduction.
RNA splicing features an intricate regulatory program that contributes to the phenotypic diversities of cells. More than 90% of human genes undergo alternative splicing, a key regulator of gene expression that generates diverse transcripts from a single protein-coding gene 1 . A splice site, a specific sequence motif located at the junction of an exon and an intron on pre-mRNA, plays a crucial role in alternative splicing. It is recognized by the spliceosome, a large RNA–protein complex that catalyzes the removal of introns, thereby guiding the splicing process 2 . Mutations could affect RNA splicing by altering splice sites, and proximal or distal cis-regulatory elements (CREs), which in turn change protein sequence and function, and result in human diseases 3 . For instance, Hutchinson–Gilford Progeria Syndrome, a rare and fatal childhood disorder characterized by accelerated aging, is caused by the activation of a cryptic splice site in the LMNA gene 4 . Therefore, recognizing variations in alternative splicing becomes an essential task for clinical diagnosis.
To overcome this challenge, computational algorithms have been developed to decipher the sequence-based splicing code and to predict the splicing effects of disease-associated genetic variations. Over the past two decades, statistical methods such as MaxEntScan 5 , and HAL 6 have been developed to identify splice sites by considering closely neighboring k-mers. Subsequently, deep learning algorithms like MMSplice 7 emerged to predict splicing effects by incorporating longer sequence contexts through the utilization of convolutional neural networks (CNNs). More recently, SpliceAI 8 , a state-of-the-art deep learning algorithm, has demonstrated significant improvements in splicing prediction compared to earlier algorithms. By leveraging complex CNN architectures, SpliceAI is capable of processing sequences of up to 10,000 base pairs. Furthermore, Pangolin 9 trained a series of models similar to SpliceAI to predict splice effects in four tissues: brain, heart, liver, and testis. In addition, other algorithms have been developed to address similar tasks in splicing prediction. For instance, CADD-Splice 10 takes outputs of multiple tools, including MMSplice and SpliceAI, as features to predict the splicing effects of genetic variants. AbSplice 11 requires the use of SpliceAI along with additional input from RNA-seq data to predict the splicing effects of genetic variations. SpliceBERT 12 , on the other hand, employs a Transformer-based model, BERT, to handle splicing-related tasks across multiple species. In practical applications, these existing algorithms have proven to be effective tools in aiding researchers in the study of disease-related gene mutations 13 , 14 . However, it is important to note that these algorithms have certain limitations in terms of accuracy and functionality when applied in real-world scenarios 15 .
A major limitation of previous methods lies in the ignorance of the tissue specificity of splicing and its clinical significance. Alternative splicing varies across cell types and tissues 16 . The brain, for example, is known to exhibit complex patterns of splicing. Variations in brain-specific alternative splicing are significantly associated with neurological disorders such as autism spectrum disorder (ASD), schizophrenia (SCZ), and bipolar disorder (BD) 17 , 18 .
For instance, aberrant splicing in CPEB4 has been reported to be highly associated with autism-like phenotype 19 . However, these alternative splicing events may not be detectable in clinically accessible tissues such as blood. Therefore, accurate prediction of splice-altering mutations in a tissue-specific manner holds significant clinical importance for genetic diagnosis. Unfortunately, most existing algorithms did not address the tissue-specificity of splicing into their model, which limits their practical application.
We present SpliceTransformer (SpTransformer), a transformer-based deep learning framework that predicts tissue-specific RNA splicing events directly from the mRNA sequence context. The success of large models in natural language processing (NLP) has proved the power of Transformer models, a deep learning architecture with multi-head attention layers. Vanilla Transformer architecture has splendid capabilities to consider sequence contexts, but requires unacceptable time and power when input comes longer. We employed attention layers specifically designed for handling long-range inputs, in order to echo the significance of considering long sequence contexts in analyzing splicing events 8 , 12 , 20 . We demonstrated the superior performance of SpTransformer compared to other deep learning models, and applied SpTransformer to accurately predict splicing effects in different tissues for various disease manifestations. Tissue-specific splicing alterations predicted by SpTransformer can be a powerful tool to guide clinical diagnosis and advance our understanding of human diseases, particularly for the less clinically accessible tissues. SpTransformer is available as an online server at http://tools.shenlab-genomics.org/tools/SpTransformer .
Tissue-specific prediction of splicing using attention-based deep learning
SpTransformer is a computational model that predicts the probability of each position in a pre-mRNA input sequence being a splice donor, splice acceptor, or neither in all and 15 different tissues simultaneously (Fig. 1 a). Previous studies have demonstrated the effectiveness of deep convolutional networks in capturing splicing patterns for general sites or specific tissues 7 , 8 , 21 . However, the multitask modeling of tissue-specific splicing poses additional challenges and has been rarely attempted. Here, we introduced the transformer architecture with multi-head attention layers to better understand the contextual features in both short- and long-range sequences (Supplementary Fig. 1 ). Additionally, we expanded the deep learning model’s background knowledge by introducing two encoders with convolution structure, and a specialized training strategy that enabled the encoders to learn from two datasets with different organizations but similar biological significance. This training strategy aimed to enable the encoders to estimate biological sequences from different hidden feature spaces, akin to a visual model that examines a human face from different perspectives. To train SpTransformer, we compiled splicing data from different sources including human adult tissue data from the GTEx database 22 , and an independent mammalian organ transcriptome dataset of four species including human, rhesus macaque, mouse, and rat 23 . We split the data by selecting subsets of chromosomes as training, and the remaining chromosomes with paralogs excluded as the testing set (see “Methods”). The relationship between the tissue classes and their corresponding detailed tissue types is presented (Supplementary Data 1 ).

a The SpTransformer model takes an only sequence as input and predicts tissue-specific splicing in 15 human tissues. The model can be used to evaluate genetic variants and predict tissue-specific splicing alterations. b Performance of 6 algorithms in splice site prediction task. Top- k accuracy is calculated by choosing a threshold to make predicted positive sites and actual splice sites have the same number, then computing the fraction of correctly predicted splice sites. PR-AUC is the area under the precision-recall curve. c Tissue-usage prediction of SpTransformer in comparison with other models. d The distribution of SpTransformer prediction score for tissue usages of splice sites in the test dataset. Tissue usages were grouped into low (<0.5) and high (≥0.5) by their original usage ratio across all samples in the same tissue types. a Created with BioRender.com, was released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
We evaluated the performance of SpTransformer on predicting all tissue splice sites in the held-out test set, and compared it against several popular methods: SpliceAI 8 , Pangolin 9 , MMSplice 7 , HAL 6 , and MaxEntScan 5 . SpTransformer achieved the highest performance in terms of both Top- k accuracy and AU-PRC, with approximately 85% top- k accuracy and 91% AU-PRC (Fig. 1 b). Most importantly, we evaluated SpTransformer prediction on different tissues in relation to the usage of splice sites across 15 human tissues. SpTransformer demonstrated a clear advantage over other tools for distinguishing splicing sites with low vs high tissue usage (Fig. 1 c, “Methods”). Specifically, the distribution of tissue-specific splicing probabilities predicted by SpTransformer aligns well with the actual usage of splice sites in each of the 15 tissues as measured by RNA-seq in the GTEx database (Fig. 1 d).
One of the enhancements of SpTransformer over previous methods is the usage of additional RNA-seq data, not only from humans but also from three other mammalian species. The similarity and homology of splicing across different mammalians have been reported in previous research 23 . Thus, we expect splicing data from diverse genomes could enhance the model’s ability to understand the tissue-specific sequence features of splicing. To investigate the real impact of including these datasets, we further conducted an ablation study using different training sets (“Methods”). SpTransformer trained with all four extra mammalian species datasets demonstrated a clear advantage over the versions trained with partial or no extra datasets (Supplementary Table 1 ). This advantage was consistently shown in the prediction of the usage of splice sites across all 15 human tissues (Supplementary Fig. 2 ). These observations underscore that the inclusion of an extra mammalian dataset enhances both the identification of splice sites and the tissue-specificity prediction. A thorough analysis of the extra mammalian datasets revealed that the primate dataset was the primary contributor to the model. We further refined the SpliceAI source code to two versions: one named SpliceAI-retrained was re-train with the same training datasets as SpTransformer, and the other one named SpliceAI-modified was modified only the output to be able to predict tissue-specificity without any change in training. We also applied other popular splicing prediction tools including MMSplice, HAL, and MaxEntScan. SpTransformer consistently outperformed them (Supplementary Table 1 and Supplementary Fig. 2 ), indicating that the superior performance is attributable not only to the training set but also to the transformer structure. Additionally, we observed that the model’s accuracy improved when it processed longer input sequences, which agreed with our expectations (Supplementary Fig. 3 ).
Regulatory code of tissue-specific splicing learned by Sp transformer
Next, we sought to understand the regulatory contexts learned by SpTransformer that contribute to the tissue-specific usage of splicing sites. As expected, splicing site tissue usage correlates with gene expression level (Fig. 2 a). This suggests that the tissue-specific splicing is, at least in part, achieved through expressional regulation, and SpTransformer implicitly learned this information by modeling tissue-specific splicing.

a Corresponding gene expression of tested splice sites in the test dataset, grouped by tissue usage of splice sites. The two-sided Fisher’s test revealed a significant association between tissue usage and gene expression of splice sites (“Low” vs “Moderate”/“High”. “Low”: 0–1 NAUC, “Moderate”: 1–20 NAUC, ''High'': over 20 NAUC. The NAUC is an estimation of a gene’s expression level, annotated by the ASCOT 66 database.). Tissue usage was not totally dominated by gene expression. b Impact of in silico mutation around intron in the GLA gene. SpTransformer considers sequence features both proximal and distal to the splice donor site. Mutagenesis weight was calculated by the decrease in the predicted strength of the splice site when that nucleotide is mutated. c Impact of in silico mutation around exons in the APBB2 gene. Several known RBP motifs were found in regions of large weight. d De novo motifs that influence the tissue-usage prediction of SpTransformer (left) and their presentations in different tissues (right). The names of similar RBP motifs, as reported by MEME tools, are marked.
It was generally accepted that the sequence context determinants, i.e., CREs, act as platforms for the recruitment of both tissue type-restricted and broadly expressed transcripts 24 , 25 . Thus, we attempted to understand the cis-regulatory sequence context learned by SpTransformer for its tissue-specific prediction performance through in silico mutagenesis. Results suggest that SpTransformer captures sequence features that are both proximal and distal to the splice site. In Fig. 2 b, we illustrate the sequence feature identified by SpTransformer using the GLA gene as an example. The GLA gene, which encodes the enzyme alpha-galactosidase A, is closely associated with Fabry disease, a rare lysosomal storage disorder. SpTransformer detected the “GT” sequence around the exon–intron junction, identifying it as a splice donor site. Additionally, SpTransformer recognized regions with relatively high mutagenesis weight at 300 nt, 400 nt, and 900 nt downstream from GLA exon 4, demonstrating its capability to detect sequence features in distal intronic regions.
Moreover, SpTransformer identified putative splicing elements that matched motifs of RNA-binding proteins (RBPs) with splicing regulatory functions reported in the corresponding tissue. For instance, the binding motif of PTBP1, a protein that plays important roles in alternative splicing in neuronal development regulation 26 , was detected in APBB2 . Alternative splicing in APBB2 results in multiple transcript variants, and polymorphisms in this gene have been associated with Alzheimer’s disease 27 (Fig. 2 c and Supplementary Fig. 4 ). To understand the sequence motifs learned by SpTransformer, we further applied motif discovery analysis with the help of the MEME toolbox 28 , and identified multiple de novo sequence motifs that influenced the model’s prediction (Fig. 2 d). Consistent with previous reports, we observed that sequences containing the consensus splicing motif AG-GT (Fig. 2 d, 1, 2) had a substantial impact on the SpTransformer model’s prediction of splicing events. This motif is the most frequently recognized sequence pattern by the spliceosomes, and could be detected in all 15 tissues by SpTransformer. Additionally, we discovered some highly specific motifs that were only present in one or two tissues (Fig. 2 d, 3–14). For example, Fig. 2 d-7 matched with the motifs of RBP DDX19B, ELAVL4, and FMR1, while Fig. 2 d-9 matched with SRSF4, according to the ATtRACT 29 database. There were also de novo motifs that were not similar to known RBP motifs.
In summary, by learning the tissue-specific splicing events, SpTransformer was able to implicitly learn the joint contribution of expression and sequence context to the tissue-specific regulatory code.
Genome-wide analysis of splicing dysregulation and human diseases
To quantify the splicing alterations of human genetic variations predicted by SpTransformer, we calculated the Δ S p l i c e score for each genetic variation and presented a graphical visualization (Fig. 3 a, “Methods”). To assess the impact of various genetic variations for splicing, we applied SpTransformer to assess the Δ S p l i c e scores of all tissue for 1,273,053 single nucleotide variants (SNVs) in the ClinVar database 30 . For instance, SpTransformer identified a cryptic splicing site with Δ S p l i c e of 0.99, which was created 24 nt upstream of the genetic mutation chr4:154569800G > T in the Fetal hemoglobin gene FGB . This mutation is known to cause Congenital Afibrinogenemia, a rare bleeding disorder featured by impairment of the blood clotting process (Fig. 3 b). Moreover, in the case of the TP53 gene mutation chr17:7675994C > A, which causes Li-Fraumeni Syndrome and predispose people to various types of cancer, SpTransformer successfully identified a cryptic splice site generated 200 nt upstream of the mutation. The splicing alterations in both cases have been experimentally validated 31 , 32 , demonstrating the accuracy of SpTransformer in predicting these distinctly different splicing alterations.

a SpTransformer is applied to evaluate the splicing effect of a single nucleotide variant by calculating an Δ S p l i c e score and matching graphical representations. b Examples of two pathogenic mutations in the ClinVar database. SpTransformer successfully predicted splicing changes even far from variants (right panel). Both cases were validated by RT-PCR in previous studies c The distribution of mutations classified by clinical significance within several intervals of Δ S p l i c e scores. As the Δ S p l i c e score increases, the ratio of pathogenic mutations becomes larger. d Distributions of Δ S p l i c e scores of all SNVs, grouped by both pathogenicity in ClinVar database and annotated variant type. The number of SNVs and the proportion of SNVs above/below the cutoff were annotated. The bar chart on the left aggregates the data by rows, while the bar chart at the top tabulates the data by columns. SNVs with alternative pathogenicity annotations (e.g., “conflicting interpretations”) were excluded from the analysis. a Created with BioRender.com, was released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
We then applied SpTransformer to make massive splicing alteration predictions on the ClinVar annotated SNVs, and assessed the performance of SpTransformer, Pangolin, and SpliceAI on distinguishing pathogenic vs benign variants. Again, SpTransformer demonstrated the best performance with AU-PRC of 0.98 (Supplementary Fig. 5 ). To gain further insights into the relationship between splicing-altering variants and pathogenic mutations, we analyzed the SpTransformer predicted Δ S p l i c e score for all ClinVar SNVs with different pathogenic labels. As expected, the proportion of pathogenic and likely pathogenic SNVs increased with higher Δ S p l i c e scores (Fig. 3 c). Specifically, as shown in Fig. 3 d, the Splice acceptor/donor sites had the most Δ S p l i c e scores close to 1, while missense and nonsense SNVs showed minimal Δ S p l i c e scores. Interestingly, we observed a bimodal distribution of Δ S p l i c e scores for intronic and synonymous SNVs in the pathogenic and likely pathogenic groups, with 57–64% of intronic (likely) pathogenic variants, and 56–58% of (likely) pathogenic synonymous variants predicted as splice altering mutations by SpTransformer. We also performed the same analysis with the retrained SpliceAI model, which showed consistency with the previous result (“Methods”, Supplementary Fig. 6 ). Identifying pathogenic variants and interpreting variants of uncertain significance (VUS) in noncoding regions and synonymous mutations has been a long-standing challenge in the field. Our analysis unveils a significant contribution of splicing alterations in intronic and synonymous pathogenic mutations, underscoring the value of applying SpTransformer in regions beyond splicing sites for diagnosis and interpretation of candidate pathogenic mutations or VUS.
Tissue-specific splicing alterations associated with disease manifestation
A variant could impact splicing universally or only in certain specific tissues. Initially, we modified Δ S p l i c e as Δ T i s s u e u s a g e scores to quantify splice alterations in each tissue. However, a drawback arose that mutations consistently affecting splicing in all tissues might receive relatively higher scores than those highly tissue-specific, making cross-tissue comparisons challenging. To address this, we derived a z -score based on the empirical distribution of selected GTEx data (Fig. 4 a and Supplementary Fig. 7 , “Methods”).

a The strategy to derive tissue specificity variants from model prediction. We created a reference set of common splicing sites to derive background distribution and calculate tissue-specific z -scores for new variants in order to make fair comparisons across tissues, and gene enrichment is calculated based on tissue-specific splice-altering SNVs. b Top five genes enriched for tissue-specific splice-altering SNVs for each of the 15 tissues as predicted by SpTransformer. The size of the bubbles represents the number of SNVs in each gene, and the color of the bubbles represents the significant level of enrichment, one-sided hypergeometric test was used for statistics. We manually examined genes associated with tissue-specific phenotypes from the HPO database and marked by a black rectangle box. c Expression pattern of top 3 genes in enrichment result of each tissue. d Proportion of pathogenic SNVs predicted as tissue-specific splice altering in different tissues. Only genes that have a p -value < 0.05 in enrichment were included. The box extends from the first quartile to the third quartile of the data, with a line at the median. The dashed line represents the median proportions of SNVs in each tissue. e Number of tissue-specific splice-altering SNVs grouped by pathogenic classifications on TTN gene in different tissues. f Genome coordinate and Tissue z -score of SNVs on a sub-region of TTN gene. SNVs are labeled with ClinVar annotation. Panel a, created with BioRender.com, was released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Next, we applied SpTransformer to all ClinVar SNVs to investigate tissue-specific splicing alterations and their association with disease clinical manifestations. We hypothesized that tissue-specific splice alterations in certain genes might be a vital contributor to the corresponding clinical manifestations. We subsequently identified the top 5 expressed genes with tissue-specific splicing alteration enrichment for each tissue (Fig. 4 b and Supplementary Fig. 8 , “Methods”). We observed a noteworthy correlation between tissue-specific splicing alterations and the corresponding disease manifestations (Fig. 4 b, box highlighted, Supplementary Data 2 ). Notably, the tissue-specificity was not predominantly influenced by gene expression (Fig. 4 c and Supplementary Fig. 9 ). In addition, the predictions of SpTransformer allowed us to evaluate the overall contribution of splicing alterations to disease across different tissues, thus we calculated the proportions of tissue-specific splice-altered SNVs in each tissue. We observed that the proportions of tissue-specific splice alterations for known pathogenic variants varied considerably across different genes and tissues (Fig. 4 d). It is worth noting that both Blood and Skin, which are considered the most clinically accessible, displayed lower proportions of tissue-specific splice alterations compared to the median average across all tissues. This observation suggests that Blood and Skin may not be suitable alternatives for estimating splicing events in other tissues.
To further examine tissue-specific splice alterations of individual genes, we conducted a detailed analysis for the heart-specific splicing-alteration enriched gene TTN as an example. SpTransformer identified over 300 tissue-specific splicing alterations of TTN in both muscle and heart, while few tissue-specific splicing alterations of TTN were identified in other tissue types (Fig. 4 e, f). The TTN gene encodes the titin protein, which is a major constituent of striated muscle proteins and is essential for sarcomere assembly, development, elasticity, and signaling 33 . In addition, we observed a large proportion of (likely) pathogenic SNVs labeled by ClinVar in the tissue-specific SNVs for TTN , suggesting a high consistency between the predicted tissue-specific splicing alterations and disease manifestation. In fact, splicing variants of the TTN gene have been reported to cause various Mendelian diseases in the heart, such as dilated cardiomyopathy (DCM), early-onset myopathy with fatal cardiomyopathy, Brugada syndrome (BrS), as well as muscle dystrophies like limb-girdle muscular dystrophy (LGMDs) and tibial muscular dystrophy (TMD) 34 , 35 , 36 , 37 , 38 , 39 , 40 . The identification of numerous tissue-specific splicing alterations in the heart further supports the capabilities of the SpTransformer algorithm.
We also probed the top genes that were enriched across different tissues in Fig. 4 b. For instance, the DMD gene, which encodes dystrophin, undergoes mutations in neuromuscular disorders such as Duchenne muscular dystrophy and Becker muscular dystrophy. These disorders are associated with cognitive impairment 41 and loss of muscle tissue 42 . Pathogenic SNVs in different DMD regions have been predicted to exhibit different tissue-specific effects (Supplementary Fig. 10 ). In a particular example, SNVs (rs1556880354 and rs2149245738) around exon 51 were predicted to be brain-specific, which aligned with previous studies that exon 51 is strongly associated with two brain-specific isoforms of DMD 43 . Lastly, the NF1 gene, which exhibits various isoforms resulting from alternative splicing, presents differential expression in muscle, blood vessels, or other cell types 44 , 45 , 46 . In our results, NF1 exhibited enrichment in muscle, blood vessels, adipose tissue, and skin (Supplementary Fig. 10 ), which is consistent with existing knowledge of NF1 mutation-induced tumorigenesis in these tissues 46 .
Together, these results suggest that SpTransformer has the capability to discern sequence features unique to tissue-specific isoforms of genes associated with disease clinical manifestations. Moreover, the SpTransformer annotation provided mechanistic insights for numerous VUS labeled in ClinVar, specifically regarding tissue-specific splicing alterations (Fig. 4 e), suggesting SpTransformer as a powerful tool to be used for genetic diagnosis and VUS interpretation purposes.
Enrichment of brain-specific splicing alterations in multiple brain disorders
Large-scale sequencing studies have been conducted on patients with brain disorders such as ASDs, SCZ, and BD, and protein-coding mutations of genes involved in neuronal activities have been mapped out 47 , 48 , 49 . To evaluate the potential clinical impact of brain-specific splicing alterations, we applied SpTransformer to predict splicing alterations in 11,986 ASD cases and 10,988 ancestry-matched controls, 24,248 SCZ cases, 3402 parent-proband trios and 97,322 controls, and 14,210 BD cases and 14,422 controls, respectively. Figure 5 a summarizes the statistics including case/control samples, and analyzed SNVs. related genes and SNVs passed the filter. We assessed the number of predicted splicing alterations across different gene regions and identified a large number of splice-altering SNVs in gene regions and mutation types beyond splice sites, e.g., intron, synonymous, nonsense and missense mutations, which underscores the potential disorder-causing role of splicing alterations outside splice sites (Fig. 5 b). In the absence of direct annotations for SNVs from the disorders dataset, we cross-validated our results with ClinVar pathogenicity annotations. Here we hypothesized that a benign SNV should not alter splicing and thus, any benign mutation predicted as splicing-altering was considered false positive. Consequently, among all filtered splicing altering SNVs ( Δ S p l i c e > 0.27 and odds ratio > 3.5) also recorded by ClinVar, the false positive rates were 7.84% (59 of 753) for ASD, 0.37% (8 of 2158) for SCZ and 4.11% (86 of 2090) for BD (Supplementary Fig. 11 ). More importantly, across all tissue types, only brain-specific splice-altering SNVs were significantly enriched in all three types of brain disorders (Fig. 5 c and Supplementary Fig. 12 ). This highlights the contribution of brain-specific splicing alterations as a common mechanism to various brain disorders.

a Statistical data for the three analyzed databases. b Splicing effect prediction for different variant types in the three brain disorder datasets: ASC, SCHEMA, and BipEx. c Enrichment of tissue-specific splicing alterations in ASD, SCZ, and BD across five tissues. A two-sided z -test for two groups was performed. The dashed line represents threshold powers for p = 0.05. d Number of tissues showing expression for genes filtered by brain-specific splicing altering SNVs in the case group. e Enriched GO term for genes in ( d ) that are expressed only in brain tissue (left) and those expressed in 11–15 tissues (right). f Network view of enriched biological processes of genes carrying brain-specific splice-altering SNVs from case group in three brain disorders. g Detailed visualization of genes enriched in GO pathway GO:0007610 “Behavior” in three brain disorders.
To gain deeper insights into the genes harboring brain-specific splice-altering SNVs, we first checked if these genes were all brain-specifically expressed. To our surprise, we observed a bimodal distribution on the cross-tissue expression of these genes (Fig. 5 d). We then carried out a gene ontology (GO) enrichment analysis and found that the genes under different expression patterns showed distinct pathway enrichment (Fig. 5 e). Those genes specifically expressed in the brain showed an explicit relation with synaptic signal, and were found to be associated with brain and mental disorders (Supplementary Fig. 13 a). On the other side, the genes expressed in 11 to 15 tissues showed functional enrichment in cytoskeleton organization (Fig. 5 g and Supplementary Fig. 13 b). For instance, DCTN1 , a gene that encodes a subunit of dynactin, has an essential role in binding microtubules and the molecular motor, and shows cytoplasmic and nuclear expression in most tissues. This gene is known to be associated with neurodegeneration including Perry syndrome 50 , and BD. We also found SHANK3 , a scaffolding protein found in excitatory synapses, which has been reported to be associated with severe cognitive deficits including language and speech disorder and ASD 51 . Furthermore, epigenetic dysregulation of SHANK3 has been linked to an increased susceptibility to ASD 52 . This analysis underscores the contribution of brain-specific splicing alteration of cytoskeleton-related genes to multiple brain disorders.
Next, we compiled all enriched pathways in a network view. As expected, the majority of biological pathways were shared across all three brain disorders. These pathways encompassed critical aspects such as brain development, synaptic signaling, neuronal system, and behavior (Fig. 5 f, Supplementary Data 3 , and Supplementary Fig. 14 ). Notably, despite sharing common pathways, the three brain disorders remained associated with different brain-specific splicing-altered genes (Fig. 5 g). We then selected “Behavior” (GO:0007610) as a study case and manually verified if the genes depicted in Fig. 5 g had been previously reported in other publications. Consequently, most of the genes identified by SpTransformer had been previously reported to be associated with corresponding disorders (Supplementary Fig. 15 and Supplementary Data 4 ). Thus, we believe that in addition to considering only gene expression, tissue-specific splicing is also crucial in clinical diagnosis.
We further investigated the consistency between genes highlighted by brain-specific splicing and other ASD-associated genomic features. For instance, a previous study by Fu et al. reported 72 ASD-associated and 373 neurodevelopmental disorder (NDD)-associated genes based on an analysis of protein-truncating variants 53 . Interestingly, the top 300 genes identified by our SpTransformer analysis of the ASD dataset showed only a small overlap with the genes reported by Fu et al. (Supplementary Fig. 16 ). Although not all those genes were investigated, our analysis did find evidence of associations between ASD and genes out of overlap. These findings underscore the importance of incorporating tissue-specific splicing patterns into the investigation of ASD genetics in order to better understand the missing inherence of this disorder.
Taken together, the findings of SpTransformer underscore the importance of investigating brain-specific splicing dysregulation as a disorder-causing mechanism for brain disorders, which holds great promise for advancing our understanding of these conditions and developing targeted therapies.
Mechanistic insights of diabetic nephropathy (DN) revealed by kidney-specific splicing alterations
Having established the SpTransformer model through GTEx, ClinVar, and multiple brain disorder large datasets, we next generated both whole exome sequencing (WES) data from blood samples and RNA-seq data of renal biopsies for an independent Chinese cohort of 95 patients with biopsy-proven DN, and applied SpTransformer to prioritize the genetic contributors to DN (Fig. 6 a). DN is a severe complication of diabetes, and the leading cause of end-stage kidney disease. While clinical evidence suggests a genetic component to DN, the genetic variations related to aberrant splicing that confer disease risk remain largely unknown. Therefore, we sought to validate the SpTransformer model using matched RNA-seq data, and to explore potential mechanisms that might contribute to disease risk through kidney-specific splicing alterations. Initially, we carried out the standard quality control procedures for WES data derived from DN patients, taking into account only those variants with a minor allele frequency (MAF) of less than 0.01 in Asia. Six thousand seven hundred forty-three SNVs were subsequently classified as splicing affecting the cutoff Δ s p l i c e > 0.27. Then, 47 of them corresponding to 46 genes were classified as kidney-specific splice-altering SNVs, which were then subjected to downstream analysis (Fig. 6 b). To validate the kidney-specific splice-altering SNVs predicted by SpTransformer, we used the matched renal tubule RNA-seq data. We only considered those SNVs that were present in different alleles among the patients in our cohort, and that had sufficient RNA-seq coverage in the region of interest. Ten unique, aberrant splicing events were validated through matched RNA-seq out of 12 predicted kidney-specific splice-altering SNVs, yielding a validation rate of 83% (Supplementary Fig. 17 ). For example, a G-to-A mutation in the CLCNKA gene created an aberrant splice site in an intron (Fig. 6 c), which subsequently resulted in a partial intron retention. The CLCNKA gene is predominantly expressed in the kidney, and the protein encoded by it is involved in transcellular chloride transport and plays a role in the maintenance of body salt and fluid balance 54 . Disruption of this gene in mice has been shown to induce nephrogenic diabetes insipidus 55 . On the other hand, a T-to-G substitution near an exon–intron junction in the BTN3A2 gene caused exon skipping (Fig. 6 d). This variant was predicted to disrupt splicing specifically in the kidney, lung, and small intestine. The BTN3A2 gene, which is expressed in multiple tissues, encodes a member of the immunoglobulin superfamily. Previous analyses have identified mutations in BTN3A2 as potential markers for immunoglobulin A nephropathy 56 .
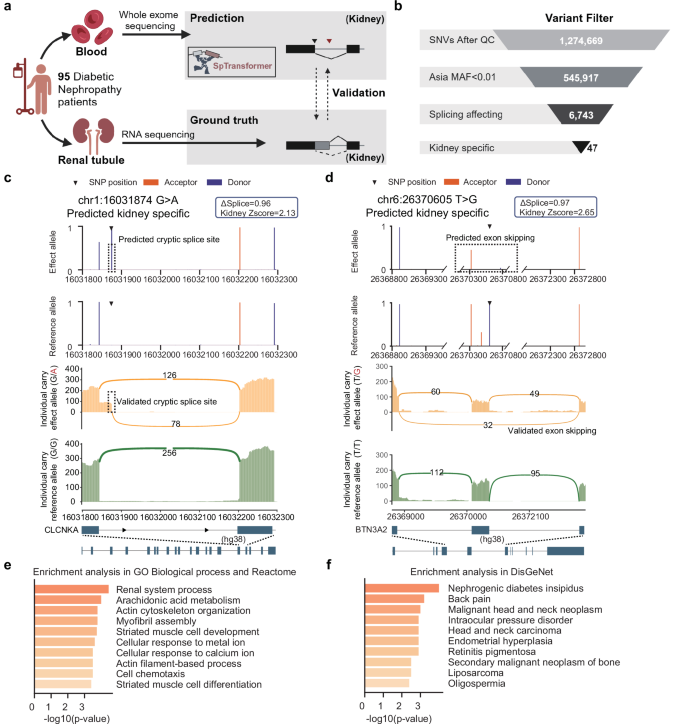
a Overview of DN patients involved and samples collected for SpTransformer prediction and RNA-seq-based validation. b Flow chart showing the filtering steps of kidney-specific splicing variants for variants called directly from WES data. c , d Examples of heterozygous variants predicted as kidney-specifically splice altering validated by matched renal tubule RNA-seq. SpTransformer prediction on WES identified variants (upper) and sashimi plot of matched RNA-seq data (lower) for CLCNKA ( c ) and BTN3A2 ( d ) gene. e Top ten GO terms enriched from genes harboring kidney-specific splicing SNVs. f Top ten terms enriched in the DisGeNet database from genes harboring kidney-specific splicing SNVs. a Created with BioRender.com, was released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
To analyze the potential mechanisms underlying the genetic risk for DN behind these genes, we performed a GO enrichment analysis and a disease terms enrichment in the DisGeNet database. The 46 genes were utilized. The most significant GO terms were reflective of kidney-specific biological processes including “renal system process” and “arachidonic acid metabolism” (Fig. 6 e), while the DisGeNet terms reflected “nephrogenic diabetes insipidus” (Fig. 6 f). Notably, arachidonic acid (AA) is a ω − 6 polyunsaturated fatty acid and its metabolites play a critical role in the pathobiology of diabetes 57 . In the kidney, prostaglandins (PG), thromboxane (Tx), and leukotrienes (LTs) are the major metabolites generated from AA 57 , 58 . An increased level of these metabolites results in inflammatory damage to the kidney 57 . Our findings in this pathway suggested that aberrant splicing may represent a potential mechanism underlying the abnormalities in AA metabolism in DN.
Together, these results support the reliability of SpTransformer prediction and enable us to explore DN candidate pathogenic variants from the perspective of splicing alterations. These findings are concordant with the known pathology of DN and highlight key genes harboring kidney-specific aberrant splicing that may contribute to renal dysfunction in DN. In summary, the application of SpTransformer helps effectively prioritize disease-associated mutations and sheds light on unresolved disease mechanisms.
Predicting RNA splicing directly from sequence data has been a long-standing challenge in the field. To address this, we have developed a novel computational framework, SpTransformer, utilizing an attention-based deep-learning neural network. SpTransformer stands out as the pioneering method to employ a transformer model for predicting RNA splicing with tissue specificity. This transformer architecture benefits SpTransformer from large-scale, cross-species datasets. In addition to predicting splicing events, SpTransformer emphasizes the tissue specificity of these events, an aspect often overlooked by most existing splicing prediction methods. This unique feature enables a more comprehensive understanding of the splicing landscape across different tissue types. SpTransformer has been successfully applied to mutation databases and disease-specific datasets, identifying tissue-specific splicing alterations and their associated disease manifestations.
Splice-altering mutations make up an essential class of known disease-causing mutations, and accurate prediction methods are crucial for interpreting VUS and pathogenic variants in clinical diagnostic tasks. While ideal scenarios would include RNA-seq profiles of diseased tissue together with the genotyping data, practically most disease-manifested tissues are not accessible or easily accessible. Nonetheless, we identified genes enriched in mutations that may alter splicing in various tissue types, which provides stronger supporting evidence for clinical interpretation of VUS pathogenicity in the relevant tissues.
As WGS becomes more widely adopted, there is an increasing need for accurate pathogenicity prediction of mutations in the noncoding regions. Through extensive analysis of the ClinVar database, we identified a significant proportion of intronic pathogenic/like pathogenic mutations that may affect splicing. Therefore, the application of splicing models, such as SpTransformer, may provide valuable information for pathogenicity prediction and interpretation of variants in the noncoding region.
The success of SpTransformer in achieving tissue specificity is attributed to the application of NLP models and large datasets. Although previous studies have demonstrated the effectiveness of deep convolutional networks in this domain, the convolution and transformer architectures we designed have several advantages. First, the tissue-specific splicing is presumably achieved through CREs as represented by sequence motifs. The attention mechanism in the transformer can help capture such CREs much more effectively. Second, transformers have demonstrated advantages in capturing distal information and can perform better at capturing CREs located far away. Third, we utilize RNA-seq data from four distinct mammalian species. This approach enables our model to discern similarities and homologies of splicing sites across species. Finally, we feed the GTEx data and evolutionary data into two convolution encoders before the transformer to extract different layers of splicing information, which supports the model with more comprehensive but structured input. Based on these considerations, our dual-encoder Transformer model, Spransformer, has exhibited a clear advantage over other state-of-the-art methods in tissue-specific splicing prediction on the GTEx dataset. To the best of our knowledge, SpTransformer is the first application of the transformer deep learning architecture that achieves remarkable performance in tissue-specific splicing prediction.
The limitations of the model come in several aspects. First, there is room for increasing the training data and including more rare splicing events. Currently, our splicing annotation is based on annotation from GTEx common splicing events, which does not account for splicing variability at the individual level. Second, the model’s inclusion of tissues is still limited. While the SpTransformer model handles approximately 15 different tissue types with high accuracy, its performance decreases as the number of multitask events in the model increases. Additionally, tissue-specific splicing is, in reality, a combined result of the splicing events of all single cells in the tissue. However, experimental technologies that measure splicing at the single-cell level are currently limited. Therefore, we envision the collection of more individualized and cell-specific splicing events can potentially enhance the deep learning model and enable more precise splicing predictions in the future.

Data representation
The input for the model is pre-mRNA sequences. The nucleotides A, C, G, and T are one-hot encoded, represented as [1,0,0,0], [0,1,0,0], [0,0,1,0], and [0,0,0,1], respectively. These encoded nucleotides are combined to form a 4 × N matrix, where N stands for the length of the sequence. [0,0,0,0] are used for padding sequences with insufficient length or to present “unknown” nucleotides in unclear regions. The model utilizes this input matrix to capture sequence features. Specifically, we denote the length of the input sequence as N = N context + N target + N context , where N target represents the length of the target region that we aim to predict, and N context represents the length of the flanking sequence to each side of the target region, which we consider as “sequence context”. For the training and testing data, each nucleotide in the target region is assigned a splice label set [ S N , S A , S D ] and a numerical tissue usage label set [ S 1 , S 2 , …, S t ]. Splice labels S A , S D , S N represent the possibility that a position is an “acceptor”, “donor” or “neither” (not a splice site), respectively. Tissue usage labels S 1 , S 2 , …, S t , where t is the number of tissues, indicate the possibility that a position is used as a splice site in a certain tissue. These labels range in [0,1]. Consequently, the model produces a matrix with the shape of (3 + t ) × N t a r g e t as its output. We applied N target = 1000, N context = 4000, and t = 15 in our work. Specifically, for an RNA sequence of length 9000 nt, SpTransformer utilized the sequence context to predict splicing sites and their corresponding usage in 15 tissues for the central 1000 nt sequence.
Data collection and labeling
We collected splicing data from various sources, including the GTEx V8 22 database and an independent large-scale RNA-seq dataset covering various species. GTEx V8 (dbGaP Accession phs000424.v8. p2) consists of 838 donors, providing 17,382 samples from 53 tissues and two cell lines. To obtain meaningful splice sites and the corresponding tissue usage ratio, we processed the exon-exon junction read counts file from the dataset for 15 representative tissue types. Initially, sequences around splice junctions in the GRCh38 reference genome were extracted. The base preceding and following each splice junction (i.e., the 5’ and 3’ ends of exons) were defined as splice sites. Only samples from the 15 selected target tissues were considered. A splice site position was labeled as “acceptor” or “donor” label if it was supported by any sample and had no conflict. The splice site at the exon start site was labeled as “acceptor”, and the splice site at the end site was labeled as “donor”. All other positions were labeled as “neither”. Subsequently, the tissue usage label was calculated for each splice site, representing the proportion of samples belonging to the tissue that contained corresponding splice junctions, i.e.,:
The tissue classes analyzed in this study included: adipose tissue, blood, blood vessels, brain, colon, heart, kidney, liver, lung, muscle, nerve, small intestine, skin, spleen, and stomach, which were frequently investigated for splicing events or had abundant samples in GTEx V8 dataset. The relationship between the class labels in SpTransformer and detailed tissue types in the GTEx was documented in Supplementary Data 1 . The SpTransformer code frameworks also support other combinations of tissue types. During the training and testing processes, any splice site with a maximum usage label of less than 0.05 across all tissue classes was excluded and re-labeled as “neither” class.
The independent RNA-seq dataset underwent similar processing steps. We utilized mammalian organ transcriptomes, including samples from humans, rhesus macaque, mice, and rats. For additional species, the genes that show orthology or paralogy to human genes in the test dataset were excluded. A splice site was identified if it was in the gene body and supported by at least one split read in each of at least two different samples.
The dataset only included tissue usage labels for 4 tissues: Brain, Heart, Liver, and Kidney. Hence, the label for each nucleotide was represented as [ S N , S A , S D , S 1 , S 2 , S 3 , and S 4 ], in accordance with the aforementioned definition. Despite the presence of additional tissue samples in the data source, such as forebrain or hindbrain, they were omitted due to their absence in all species. Since there were directly accessible RNA-seq data, the tissue usage label was estimated by splice site strength calculated with SpliSER 59 . The dataset was partitioned following the same strategy as the GTEx dataset. The part of the training data was considered an extension of the training dataset, while the test data segment remained unused.
We chose sequences from chromosomes 2, 4, 6, 8, 10–22, X , and Y to create the training dataset. Sequences belonging to chromosomes 1, 3, 5, 7, and 9 were compiled as the testing dataset. To ensure the test data was not exposed in training data, we cross-referenced the sequence strands with the Ensemble database (release 110) 60 . We excluded gene sequences that have paralogs from the testing dataset. Despite splitting the two datasets independently, we made sure that there was no overlap between the training and testing data after the steps to split data by chromosomes and paralogs.
To compile the datasets, the pre-mRNA sequences of each gene were extracted. For genes with multiple transcripts, the extracted sequence began from the most upstream site observed across all transcripts and ended at the most downstream site observed across all transcripts. Subsequently, each sequence was divided into blocks of length 1000 nt. Blocks that did not contain any splice sites were discarded. For each remaining block, the flanking sequence with 4000 nt + 4000 nt and the corresponding 1000 nt label was packaged as a single training (testing) data entry.
SpTransformer model structure
SpTransformer is a large deep neural network model that consists of two main modules: an encoder module and a transformer module. The encoder module incorporates a series of residual networks, known as ResBlock, with inspiration originating from classic ResNet structure 61 in computer vision tasks. Previous studies have shown the effectiveness of convolution-based methods in capturing sequence or “motif” features in DNA/RNA sequences 7 , 62 . However, as the depth of the network increases, a brutal stack of convolution layers may lead to a rapid decrease in the network’s accuracy due to the vanishing or exploding gradient problem 61 . A residual block is composed of a series of convolution layers, interspersed with several batch-normalization layers and RELU activation function layers. Stacking these residual blocks can help mitigate the gradient issue and enhance the model’s ability to handle complex features 61 .
The second component of our model mainly consists of a Sinkhorn Transformer module and a multi-layer perception module. Transformers have become a fundamental component in the field of modern NLP 63 . Their “Attention” mechanism efficiently solved sequence-based tasks such as the Named Entity Recognition problem, which is similar to our challenge of recognizing splicing sites based on RNA sequences. The Sinkhorn Transformer 64 is a variant of the transformer model that is designed to handle sequences of considerable length (8192 nt in our model).
The architecture of our model is shown in Supplementary Fig. 1 . The input is an RNA sequence of length N = N context + N target + N context , where N target denotes the length of the target sequence, and N context represents the sequence context of the target region ( N context = 4000 in our pipeline). The parameter L dictates the number of channels in each convolution layer, with L 1 = 192 and L 2 = 64 used in this study. The convolution layer in ResBlocks is characterized by parameters L , W , and D , which represent the number of channels, kernel width, and dilation, respectively. Following the calculation of the encoder module, a concatenation, and a truncation operation are applied to ensure that the input to the attention module does not exceed a length of 8192. The Sinkhorn Transformer module has 256 channels, 8 attention heads (including two local attention heads) per layer, and eight layers of depth. The final output is a ( N × 3) shaped matrix and a ( N × 15) shaped matrix, representing splice site prediction and tissue usage prediction, respectively.
The hyperparameters were selected by multiple processes: 1) N target and N context : the determination of these parameters was influenced by the model’s architecture. While our goal was to allow the model to process long sequences, practical constraints—such as GPU memory limitations—necessitated a compromise. In that case, we set N target = 1000, N context = 4000 so that each target nucleotide has at least 4000 + 4000 available context information, taken considering that 8000 nt context is enough for the model (Supplementary Fig. 3 ). 2) The dimension of encoder layers was gained by grid-search in {32, 64, 128, 256}. Multiple hyperparameters of the transformer module were also tried, including: layers of transformer = {4, 6, 8, 10}, and number of attention heads = {6, 8, 10}. During this process, batch size = 12 and learning rate = 0.001 were used in order to keep consistent with SpliceAI. 3) Other parameters was selected from: batch size = {6, 12, 18, 24}, learning rate = {0.01, 0.005, 0.001, 0.0001}, decay factor of learning rate = {0.9, 0.7, 0.5}. Those options were established in reference to previous publications, suggestions from Transformer models, and the memory limitations of GPUs. From all those options, the combination with the best performance on the validation dataset was subsequently used.
Further details of input and output have been provided in the “Data representation” section. Different measurement was applied to the output scores. For splice site prediction, we applied a Softmax activation function to produce probability prediction of “Acceptor”, “Donor”, and “Neither”. For tissue usage prediction, we applied a sigmoid activation for each tissue type.
Model training and testing
SpTransformer underwent two stages of training. In the first stage, two convolution-based encoder modules with identical structures were trained separately. One module learned weights from the GTEx training dataset, while the other module learned weights from the excessive training dataset, due to the fact that the two datasets have different label formats. To accommodate the different data formats of the two datasets, temporary convolution layers with 1 × 1 kernel size and particular output channels (18 and 7, respectively) were appended to each convolution module to align the outputs with the labels. The output of the temporary layers was used directly for loss calculation and back propagation. The two modules were assigned 128 and 64 channels (i.e., the parameter “L” in Supplementary Fig. 1 ), respectively, based on the difference in tissue numbers. In the second stage, the temporary layers were removed. The two modules with frozen parameters were directly integrated into a 192-channel convolution network, as shown in Supplementary Fig. 1 . The whole network was then trained on the GTEx training dataset to get the final model. This approach enabled SpTransformer to learn from multiple datasets with similar biological meanings but different data formats, minimizing the need for extensive coding or conversion when differently sourced data was received. Despite both datasets being under the sequence model, the diverse splicing representations and distinct data content encourage the deep model to comprehend latent sequence features from various aspects, akin to a visual model examining a human face in multiple ways. The strategy improved the model’s performance compared to only using one dataset, demonstrating the potential to integrate diverse bioinformatics data for a single task.
Special loss functions are used in the backpropagation of deep learning. In the first stage, the loss function is:
Each sequence in the training dataset is a contiguous nucleotide sequence of length n . The i -th position has a splicing label A i and a tissue-usage label B t i for T different tissues. For this position, the model outputs s i for splice site prediction, and outputs u t i for tissue-usage prediction. For splicing prediction, we compute the categorical cross-entropy loss, as it is a multi-class classification task. In contrast, for tissue-usage prediction, which is a multi-label classification task (meaning one sample can belong to multiple classes), we calculate the Binary Cross-Entropy loss. We apply mean reduction for the two loss functions.
The loss function above is sufficient for the encoders to learn basic sequence patterns. However, as the number of supported tissues increases, new challenges emerge. First, models struggle to learn the features of different tissues in a balanced manner during the training process, occasionally demonstrating superior predictive performance for specific tissues only. Second, there is a relative scarcity of samples with strong tissue specificity compared to those with weak tissue specificity. This imbalance leads to models tending to produce similar tissue usage scores for splice sites. During the second stage, we apply a special loss function, in order to persuade the transformer module to overcome these difficulties:
The formula calculates the weighted geometric mean for loss of tissue usage. The idea is motivated by previous computer vision research 65 . We use this method to encourage the model to balance the performance on multiple tissues and pay attention to those tissues that harder to classify. For the i -th position of the sequence, a weight w i was multiplied to encourage the model to pay more attention to splice sites with stronger tissue specificity. w i related with the variance of tissue-usage labels of i -th position.
The model was trained for 12 epochs in each stage. Adam optimizer was used to minimize the combined loss. The learning rate was set to 0.005, and was multiplied by 0.7 after every epoch.
Model performance evaluation and ablation test
We evaluated the performance of SpTransformer on two tasks using the compiled test dataset: 1) splice site prediction in long sequences: the model took each pre-mRNA sequence as input and identified every splice acceptor and donor within a target region of 1000 nt. Given that most positions in the sequences are not splice sites, we computed the top- k accuracy and the area under the precision-recall curve (AU-PRC) for splice site prediction. The top- k accuracy was defined as follows: if a sequence has k positive positions that truly belong to the class, a threshold is selected so that exactly k positions are predicted to be positive. The fraction of these k predicted positions that truly belong to the class is reported as the top- k accuracy. We calculated the top- k accuracy and AU-PRC value for the acceptor and donor classes separately, and reported the average performance of the two classes. 2) Tissue usage level prediction: the model was tasked with predicting the usage level in each of the 15 tissue classes for each position of the sequence. Given the absence of a widely accepted “tissue usage” protocol, we divided all splice sites in the test dataset based on their tissue usage label. Since most tissue usage labels were close to (or equal to) 0 or 1, we defined a tissue usage label greater than 0.5 as “high usage”, while the remaining sites were classified as “low usage.” The usage was set to 0 if a position was not a splice site. The model was then tasked to classify the usage of each position in the test dataset. Furthermore, positions that did not pass the top- k threshold in task 1 were forcibly masked as negative in the prediction result. We calculated AU-PRC for each tissue class. According to the two tasks outlined above, we conducted a comparative analysis, including an ablation test to highlight the advantages of our method. Firstly, we prepared three different versions of the SpTransformer model: SpTransformer-noextra, which was trained only on the GTEx training dataset; SpTransformer-extra1, which used an extra training dataset of two species, human and macaque; and SpTransformer, which used the full training dataset of four mammalian species. All versions were trained with the same configuration.
Secondly, we applied the published version of SpliceAI to the tasks for comparison. Furthermore, we prepared a similar CNN-based network, named as SpliceAI-modified. The key difference was a specific alteration: the output channel number of the final convolution layer was adjusted from 3 to 18. This modification enabled the model to predict tissue usage at each splice site. The modification was a permissible adjustment within the conventional design framework of CNN. This network was trained on the GTEx dataset with the same hyperparameter and loss functions as the first stage of SpTransformer. We also retrained SpliceAI on our training dataset using the same hyperparameters as the original version, named SpliceAI-retrained. We then included the published version of Pangolin. Pangolin supports the prediction of four tissues (brain, heart, liver, and testis) and does not distinguish between acceptor and donor. Thus, the Brain, Heart, and Liver models of Pangolin were used. The maximum splice scores of them were used in task 1. The tissue scores of them were used in task 2. It is worth noting that SpliceAI-modified has a remarkably similar structure to Pangolin, despite the differences in their last output layers. SpliceAI-modified predicts splicing effects across 15 tissues using a single model, whereas Pangolin utilizes four distinct models, each dedicated to a specific tissue.
Finally, we adapted the task for earlier methods that were not entirely compatible with our tasks. MMSplice, HAL, and MaxEntScan were designed for classifying a single position with short flanking sequences. We modified the input format to enable them to predict each position of the long sequences individually. For each position, the lengths of input sequence were carefully selected based on the recommendations provided by their respective publications. Since HAL does not score 3’ splice sites, we excluded the “Acceptor” class when evaluating HAL. The “MMSplice_MTSplice” tool is a combined model where “MMSplice” predicts splice sites and “MTSplice” predicts tissue-specific usage of those splice sites. During testing, we evaluated MMSplice in two different scenarios: the full dataset, as well as a simpler task where it was restricted to predicting positions within a 20 nucleotide range of each splice site, rather than the whole sequences. The performance on the simpler task was marked as “MMSplice-short”. MTSplice was able to predict usage scores in 56 detailed tissue types. We took the maximum score of corresponding types as the prediction of 15 classes in our dataset. Since “MMSplice-short” exhibited an advantage against “MMSplice”, MTSplice was also tested on the restricted task.
We further investigated the importance of context in flanking RNA sequences of splice sites. All splice sites in chromosome 1 were extracted from the GTEx test dataset. Several test routines were performed after masking flanking sequences over certain values of the range. AU-ROC was calculated for each test routine in the same way as performance testing before (Supplementary Fig. 3 ).
Estimate sequence weights by in-silico mutagenesis
To identify important regions within the input sequences, we performed a procedure referred to as “in silico mutagenesis”. The “mutagenesis weight” of a nucleotide with respect to a splice site is defined as follows: Let s ref denote the splice (or tissue usage) score of the target splice site. The score is recalculated by replacing the nucleotide under consideration with A, C, G, and T. Let these scores be denoted by s A , s C , s G , and s T , respectively. The mutagenesis weight of the nucleotide is estimated as:
Under our hypothesis, a larger mutagenesis value indicates that “the splice site is more likely to be lost if the position is mutated”. We calculated the difference between the maximum and mean tissue usage values across all 15 tissues for each splice site in the GTEx dataset. Those with the top 1% difference value were selected, including 2754 splice donors and 2670 splice acceptors. For each selected splice site, in silico mutagenesis was performed for the sequence from 80 nt upstream to 20 nt downstream of the site. For each sequence, let max s r e f denote the maximum s ref value among all the 15 tissues. For each tissue and each sequence, all subregions with 8 nt length and with average mutagenesis weight greater than m a x sref were extracted. The motif analysis tool XSTREME 28 was utilized to enrich motifs from those extracted subregions. Multiple associating motifs were enriched for each tissue through the outlined methodology. Despite the different PWM matrices, the motifs were treated as the same term if their IUPAC codes (given by XSTREME) have a Levenshtein Distance not greater than 1.
Quantifying splicing change resulting from variants
In order to quantify the splicing alterations caused by SNVs, we calculated the difference in scores between the original sequences and the alternated sequences. Firstly, we predicted the 2 R + 1 length of the reference sequence surrounding the mutation ( R represents the length of flanking nucleotides on each side, and was set to 100 by default in our analysis) using SpTransformer. We then used the alternative sequence for prediction, resulting in prediction scores for each position of the sequence. Regardless of the “not a splice site” class, scores for each class were represented by vectors in the shape of (2 R + 1) × 1. For reference and alternated sequence, there were scores named R e f acceptor , Ref donor , A l t acceptor , A l t donor , R e f brain , R e f heart , A l t brain , A l t heart , …, for two splice site types and 15 tissue types. We evaluated the following quantities, using brain tissue as an example:
Δ s c o r e for other tissues was calculated in the same way, where abs() refers to the function to calculate the absolute value for each position, and max() is the function to find the max value in the vector. We defined Δ S p l i c e as the max value between Δ A c c e p t o r and Δ D o n o r to quantify the effect of splice alteration caused by a variant:
The Δ s c o r e for each tissue, for example, Δ B r a i n , was processed to quantify the change in tissue usage.
Quantify tissue-specificity by tissue z -score
To quantify tissue-specificity, we designed a z -score. We created a reference mutation set with GTEx SNVs data. Upon checking the GTEx Genotype calls vcf file, we identified a total of 734,509,842 variants. We selected SNVs that met the following conditions: 1) For each tissue, there should be at least one available RNA-seq data from an individual carrying the SNV. 2) For each tissue, there should be at least one available RNA-seq data from an individual not carrying the SNV. 3) Within a 100nt range of the SNV location, a significant change in splice site usage, either increase or decrease, should be observable across all tissues when comparing RNA-seq data between individuals with and without the SNV. The “low” and “high” were under the same definition as those used in the training dataset. Finally, we filtered out 27,843 mutations that cause splice alterations in all tissue classes. These mutations were expected to have minimal impact on tissue specificity, and their Δ s c o r e was used to build a reference distribution hereafter. SpTransformer predicted the Δ S p l i c e score and tissue Δ s c o r e s for each mutation. A tissue z -score was then calculated based on the following formula. Here we use adipose as an example.
Here we denote μ as the average values, σ as the standard error, and Z i representing the adipose tissue z -score for the i -th mutation.
The distribution of the tissue z -score for each tissue was defined as the reference distribution mentioned above (Supplementary Fig. 7 ). For any new SNVs analyzed, we similarly calculated its z -score using previously calculated μ tissue and σ tissue . For each tissue, we consider any real SNVs with a tissue z -score greater than X % of the reference distribution to be tissue-specific. In all analyses in this work, X = 99 was used by default.
Quantifying gene expression level by ASCOT database
We employed the normalized area under the curve (NAUC) as a measure of gene expression levels. The introduction of NAUC by ASCOT 66 enables users to explore gene expression in a wide range of cell types and tissues. The dataset was derived from the GTEx project, which included 9662 human RNA-Seq samples across 53 tissues. In our study, we calculated the average NAUC value across the corresponding tissue types for each of the 15 tissue classes. The correlation between the tissue classes and their corresponding detailed tissue types is presented in Supplementary Data 1 . We classified gene expression into “Low” (0–1 NAUC), “Moderate” (1–20 NAUC), and “High” (over 20 NAUC) according to the recommended standard. This classification was used during analysis to exclude genes with “Low” expression in the tissues of interest from our investigation. All the gene expression values presented in the figures were based on the NAUC values.
Finding splicing altering SNVs in the ClinVar database
We utilized SpTransformer to predict splicing altering variants in the ClinVar database (the 20220917 version with hg38 genome annotation). We analyzed a total of 1,273,053 SNVs. These SNVs were annotated using SnpEff 67 and only variants with consequence annotations “splice acceptor variant”, “splice donor variant”, “intron variant”, “synonymous variant”, “missense variant”, and “nonsense” were considered in all analyses. We employed Δ S p l i c e to distinguish variants of different pathogenicity (Fig. 3 a). The SNVs were categorized based on their consequence annotations (e.g., “Intron”, “Synonymous”, etc.) and clinical significance labels (e.g., “Benign”, “Pathogenic”, etc.). SNVs with ambiguous labels such as “Conflicting interpretations of pathogenicity” or “Likely risk allele” were excluded.
We further utilized SNVs from the ClinVar dataset to establish a score threshold for SpTransformer (Supplementary Fig. 5 ). The “Strict” panel presents the performance of our SpTransformer model in distinguishing between pathogenic and benign mutations, while the “Soft” panel presents the performance in distinguishing between pathogenic/likely pathogenic/uncertain mutations and benign/likely benign mutations.
The classification cutoff was determined by considering the F1-score, recall, and precision in these tasks. We employed a cutoff value of 0.27, ensuring that the model demonstrated equal precision and recall in the “Strict” task, based on the following three considerations: 1) to specifically investigate splicing events. 2) The F1-score showed only slight differences across a wide range of delta splice values, while recall dropped off more significantly (Supplementary Fig. 5 c). 3) In a clinical setting, we had a preference for higher recall over higher precision, in order to identify critical variants. This cutoff value allowed us to discern variants that impact splicing. When analyzing Figs. 4 and 5 , the cutoff value served as a filter to identify mutations that have the highest likelihood of being implicated in human disease through their impact on splicing. We also provided other cutoffs for users. E.g., Δ s p l i c e = 0.54 for the max F1-score, and Δ s p l i c e = 0.09 for distinguishing “likely pathogenic” and “likely benign” mutations. Similarly, a cutoff = 0.32 was chosen for the SpliceAI-retrained model, to reproduce SpTransformer’s result as a comparison (Supplementary Fig. 6 ).
Enriching genes in tissue-specifically splice-altering SNVs
To identify genes in ClinVar associated with tissue-specific splicing, we performed an enrichment pipeline for SNVs. This pipeline incorporated three filters to identify tissue-specific splicing SNVs: Δ S p l i c e score = 0.27, tissue z -score > 99%, and ASCOT gene expression NAUC > 1. Subsequently, we conducted a hypergeometric test on the genes that contained those predicted SNVs. To validate the association between gene-tissue disease, we referred to the Human Phenotype Ontology (HPO) database 68 . For each gene enriched with splicing alterations SNVs, we examined HPO terms for tissue-related phenotypes in the corresponding Mendelian disorders.
Process and analysis of three different brain disorders
We collected variants related to three brain disorders: ASD, SCZ, and BD from three different public databases ASC, SCHEMA, and BipEx, respectively. The ASD data was obtained from the analysis of de novo variants called in family-based data consisting of 6430 probands with ASD and 2179 unaffected siblings, as well as rare variants called in 5556 ASD cases and 8809 ancestry-matched controls. The SCZ data was obtained from the GATK analysis of WES of 24,248 cases and 97,322 controls, as well as de novo mutations from 3402 parent-proband trios. The BP data was obtained from variants called from WES of 14,210 cases and 14,422 controls, post-quality control. Due to limited raw sample data, we were unable to segregate mutations into case and control groups using the GATK pipelines and calculate p -values. Instead, we calculated the odds ratio (OR) using the allele count and allele frequency of SNVs in the case and control samples. SNVs with an OR greater than 3.5 were classified as case SNVs. Following this pipeline, we employed SpTransformer to predict the splicing effect. After prediction, we applied a series of filters: 1) SNVs with Δ S p l i c e greater than 0.27 (the cutoff determined in Supplementary Fig. 5 ) were considered splice-affecting SNVs. 2) SNVs with brain tissue z -score above 99% and the corresponding genes had moderate expression (NAUC > 1) were finally categorized as brain-specific splicing variants. The two-proportion z -test used in Fig. 5 c is a statistical test designed to determine whether the proportions of categories in two group variables significantly differ from each other. In this study, we employed this test to examine whether there exists a larger proportion of brain-specific splicing variants among case SNVs that are related to splicing. Specifically, Group A consisted of all SNVs with O R > 3.5 and Δ S p l i c e ≥ 0.27. while Group B included all SNVs with O R ≤ 3.5 and Δ S p l i c e ≥ 0.27.
We sorted all filtered SNVs by brain tissue z -score and selected the top 300 related genes for each disorder. We applied the online tool Metascape 69 for GO terms enrichment analysis, disease–gene association enrichment analysis, and corresponding network figures. We categorized tissue-specific expression patterns based on the ASCOT NAUC values across 15 tissue classes. A gene was designated as having ’brain-specific expression’ if its NAUC value exceeded 1 solely in brain tissue. Similarly, if a gene’s NAUC value exceeded 1 in any other tissue, it was classified as expressed in that specific tissue. We further manually verified whether each gene listed in Fig. 5 g had been previously reported to be associated with the respective disorders. Genes were classified as “previously reported” if any publication in PubMed explicitly stated in the abstract, results, or discussion that the gene is associated with the disorders.
DN variant enrichment and RNA-seq validation
We processed WES data from DN patients for downstream analysis. Genomic DNA isolated from whole blood was used for genotyping. Ninety-five DN patients were genotyped using Agilent SureSelect Human All Exon V6 kit (Agilent Technologies) and sequenced on an Illumina Hiseq3000 with Hiseq3000SBS&Clusterkit. The human kidney tissues used were obtained from DN patients with renal biopsy-proven, sourced from Nanjing Glomerulonephritis Registry, National Clinical Research Center for Kidney Diseases, Jinling Hospital, Nanjing, China. RNA sequencing was performed on micro-dissected tubulointerstitial tissue from 95 patients diagnosed with DN. Total RNA was extracted using an RNAeasy Micro kit (Cat#74,004, QIAGEN Science) according to the manufacturer’s instructions. RNA quality was assessed with the Agilent Bioanalyzer 2100. Only samples with RIN scores above seven were used for cDNA production. Qualified total RNA was further purified by RNAClean XP Kit (Cat A63987, Beckman Coulter, Inc. Kraemer Boulevard Brea, CA, USA) and RNase-Free DNase Set (Cat#79254, QIAGEN, GmBH, Germany). Strand-specific, polyA-enriched RNA-seq libraries were generated using the VAHTS Stranded mRNA-seq Library Prep Kit for Illumina. FastQC (v0.11.9) was used for quality control. STAR (v2.7.8a) 70 was used for alignment against the human Grch38 assembly. Transcripts of low quality were filtered out, and the read counts were normalized to transcripts per million (TPM) within each sample. These values were then log2 transformed (log2 (TPM + 1)). To identify kidney-specific splice-altering SNVs, we performed the following steps: 1) applied the GATK variant caller and standard quality control procedures; 2) retained the SNVs with an MAF of less than 0.01 in the Asian population; and 3) retained the SNVs with an Δ S p l i c e greater than 0.27, a kidney tissue z -score above 95% and the corresponding genes had expression NAUC score > 1 in the kidney. These procedures resulted in 47 kidney-specific splice-altering SNVs corresponding to 46 genes expressed in the kidney tissue (Fig. 6 b). We validated these retained SNVs using RNA-seq data, excluding those without sufficient coverage at the mutation site and adjacent exon regions. Following this pipeline, 12 SNVs corresponding to 12 genes were validated. Each SNV was manually checked in IGV browser 71 and visualized with the R package ggsashimi 72 .
Statistical information
Statistical tests are described in the figure captions, main text, and corresponding “Method” sections. To summarize, we used a one-tailed hypergeometric test to enrich tissue-specific splicing-altering SNVs in genes from the ClinVar database. We used a two-sided z -test for two groups to measure the enrichment of tissue-specific SNVs in the three brain disorder datasets. For tissue z -scores defined in the main text, we use sentences like “99% significance” to represent tissue z -scores that are greater than 99% of mutations in the reference distribution. The GO term enrichment analysis (related to Figs. 5 e, f and 6 e, f) was calculated by Metascape based on the hypergeometric test and Benjamini–Hochberg p -value correction algorithm.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All public data and tools utilized in the analysis are referenced in the main text or the Supplementary Materials . Detailed information on the data sets, learning algorithms, and statistical analyses can be found in the Supplementary Materials . The utilized public datasets are listed below: GTEx exon–exon junctions data are available from the GTEx analysis V8 (dbGaP Accession phs000424.v8.p2) [ https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v8.p2 ]; the excessive RNA-seq data were published in ref. 23 ; The hg38 genome annotation and gene paralogs data [ www.ensembl.org ]; the ClinVar database (2022-09-17, hg38 version) [ www.ncbi.nlm.nih.gov/clinvar/ ]; the ASCOT gene expression data were published in ref. 66 ; the Autism Sequencing Consortium (ASC) database was published in ref. 47 [asc.broadinstitute.org/]; the SCZ exome meta-analysis consortium (SCHEMA) data was published in ref. 48 [schema.broadinstitute.org/]; the Bipolar Exomes (BipEx) data was published in ref. 49 [bipex.broadinstitute.org/]; and the HPO database 73 [ https://hpo.jax.org/app/ ]. The transcriptome profiling of the tubulointerstitial compartment from patients with diabetic kidney disease is available under accession number OMIX006095 . Access to data generated in this study for research purposes is available upon request from the corresponding author. Source data are provided with this paper.
Code availability
Code about the SpTransformer program 74 is available under an Apache-2.0 license on GitHub at https://github.com/ShenLab-Genomics/SpliceTransformer .
Tazi, J., Bakkour, N. & Stamm, S. Alternative splicing and disease. Biochim. Biophys. Acta 1792 , 14–26 (2009).
Article CAS PubMed Google Scholar
Wang, Z. & Burge, C. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14 , 802–13 (2008).
Article CAS PubMed PubMed Central Google Scholar
Pagani, F. & Baralle, F. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 5 , 389–96 (2004).
Ahmed, M. S., Ikram, S., Bibi, N. & Mir, A. Hutchinson–Gilford progeria syndrome: a premature aging disease. Mol. Neurobiol. 55 , 4417–4427 (2018).
CAS PubMed Google Scholar
Yeo, G. & Burge, C. Maximum entropy modeling of short sequence motifs with applications to rna splicing signals. J. Comput. Biol. 11 , 377–94 (2004).
Rosenberg, A., Patwardhan, R., Shendure, J. & Seelig, G. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 163 , 698–711 (2015).
Cheng, J. et al. Mmsplice: modular modeling improves the predictions of genetic variant effects on splicing. Genome Biol. 20 , 48 (2019).
Article PubMed PubMed Central Google Scholar
Jaganathan, K. et al. Predicting splicing from primary sequence with deep learning. Cell 176 , 535–548.e24 (2019).
Article PubMed Google Scholar
Zeng, T. & Li, Y. Predicting rna splicing from dna sequence using pangolin. Genome Biol. 23 , 103 (2022).
Rentzsch, P., Schubach, M., Shendure, J. & Kircher, M. Cadd-splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 13 , 1–12 (2021).
Article Google Scholar
Wagner, N. et al. Aberrant splicing prediction across human tissues. Nat. Genet. 55 , 1–10 (2023).
Chen, K. et al. Self-supervised learning on millions of primary RNA sequences from 72 vertebrates improves sequence-based RNA splicing prediction. Brief. Bioinforma. 25 , bbae163 (2024).
Wai, H. et al. Blood RNA analysis can increase clinical diagnostic rate and resolve variants of uncertain significance. Genet. Med. 22 , 1005–1014 (2020).
Richter, F. et al. A deep intronic pkhd1 variant identified by spliceAI in a deceased neonate with autosomal recessive polycystic kidney disease. Am. J. Kidney Dis. 83 , 829–833 (2024).
Yépez, V. A. et al. Clinical implementation of rna sequencing for mendelian disease diagnostics. Genome Med. 14 , 38 (2022).
Tao, Y., Zhang, Q., Wang, H., Yang, X. & Mu, H. Alternative splicing and related RNA binding proteins in human health and disease. Signal Transduct. Target. Ther. 9 , 26 (2024).
Porter, R., Jaamour, F. & Iwase, S. Neuron-specific alternative splicing of transcriptional machineries: implications for neurodevelopmental disorders. Mol. Cell. Neurosci. 87 , 35–45 (2017).
Gandal, M. et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362 , eaat8127 (2018).
Article ADS CAS PubMed PubMed Central Google Scholar
Parras, A. et al. Autism-like phenotype and risk gene mrna deadenylation by cpeb4 mis-splicing. Nature 560 , 441–446 (2018).
Margasyuk, S. et al. Rna in situ conformation sequencing reveals novel long-range rna structures with impact on splicing. RNA 29 , rna.079508.122 (2023).
Xiong, H. Y. et al. The human splicing code reveals new insights into the genetic determinants of disease. Science 347 , 1254806 (2015).
Article MathSciNet PubMed Google Scholar
Consortium, T. G. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science 369 , 1318–1330 (2020).
Cardoso-Moreira, M. et al. Gene expression across mammalian organ development. Nature 571 , 505–509 (2019).
Smith, A., Sumazin, P. & Zhang, M. Tissue-specific regulatory elements in mammalian promoters. Mol. Syst. Biol. 3 , 73 (2007).
Das, D. et al. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 35 , 4845–57 (2007).
Liu, H.-L. et al. The role of rna splicing factor ptbp1 in neuronal development. Biochim. Biophys. Acta Mol. Cell Res . 1870 , 119506 (2023).
Golanska, E. et al. Analysis of APBB2 gene polymorphisms in sporadic Alzheimer’s disease. Neurosci. Lett. 447 , 164–166 (2008).
Grant, C. E. & Bailey, T. L. Xstreme: comprehensive motif analysis of biological sequence datasets. Preprint at https://doi.org/10.1101/2021.09.02.458722 (2021).
Giudice, G., Sánchez-Cabo, F., Torroja, C. & Lara-Pezzi, E. Attract—a database of rna-binding proteins and associated motifs. Database 2016 , baw035 (2016).
Landrum, M. J. et al. Clinvar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42 , D980–5 (2014).
Varley, J. M. et al. Characterization of germline tp53 splicing mutations and their genetic and functional analysis. Oncogene 20 , 2647–2654 (2001).
Spena, S. et al. Congenital afibrinogenemia: first identification of splicing mutations in the fibrinogen bbeta-chain gene causing activation of cryptic splice sites. Blood 100 , 4478–84 (2002).
Trinick, J., Knight, P. & Whiting, A. Purification and properties of native titin. J. Mol. Biol. 180 , 331–56 (1984).
Zheng, W. et al. Identification of a novel mutation in the titin gene in a chinese family with limb-girdle muscular dystrophy 2j. Mol. Neurobiol. 53 , 5097–102 (2016).
Khan, A. et al. Homozygous missense variant in the ttn gene causing autosomal recessive limb-girdle muscular dystrophy type 10. BMC Med. Genet. 20 , 166 (2019).
Hackman, P. et al. Tibial muscular dystrophy is a titinopathy caused by mutations in ttn, the gene encoding the giant skeletal-muscle protein titin. Am. J. Hum. Genet. 71 , 492–500 (2002).
Hackman, P. et al. Truncating mutations in C-terminal titin may cause more severe tibial muscular dystrophy (tmd). Neuromuscul. Disord. 18 , 922–8 (2008).
Pfeffer, G. et al. Titin founder mutation is a common cause of myofibrillar myopathy with early respiratory failure. J. Neurol. Neurosurg. Psychiatry 85 , 331–8 (2014).
Carmignac, V. et al. C-terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann. Neurol. 61 , 340–51 (2007).
Wang, L. L. et al. Genetic profile and clinical characteristics of Brugada syndrome in the Chinese population. J. Cardiovasc Dev. Dis. 9 , 369 (2022).
CAS PubMed PubMed Central Google Scholar
Bresolin, N. et al. Cognitive impairment in duchenne muscular dystrophy. Neuromuscul. Disord. 4 , 359–369 (1994).
Wilson, K. et al. Duchenne and becker muscular dystrophies: a review of animal models, clinical end points, and biomarker quantification. Toxicol. Pathol. 45 , 961–976 (2017).
Doisy, M. et al. Networking to optimize dmd exon 53 skipping in the brain of mdx52 mouse model. Biomedicines 11 , 3243 (2023).
Trovó-Marqui, A. & Tajara, E. Neurofibromin: a general outlook. Clin. Genet. 70 , 1–13 (2006).
Gutmann, D., Cole, J. & Collins, F. Modulation of neurofibromatosis type 1 (nf1) gene expression during in vitro myoblast differentiation. J. Neurosci. Res. 37 , 398–405 (1994).
Staser, K., Yang, F.-C. & Clapp, D. Mast cells and the neurofibroma microenvironment. Blood 116 , 157–64 (2010).
Satterstrom, F. K. et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180 , 568–584.e23 (2020).
Singh, T. et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604 , 509–516 (2022).
Palmer, D. S. et al. Exome sequencing in bipolar disorder identifies akap11 as a risk gene shared with schizophrenia. Nat. Genet. 54 , 541–547 (2022).
Konno, T. et al. Dctn1-related neurodegeneration: Perry syndrome and beyond. Parkinsonism Relat. Disord. 41 , 14–24 (2017).
Durand, C. M. et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39 , 25–27 (2007).
Zhu, L. et al. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum. Mol. Genet. 23 , 1563–1578 (2014).
Fu, J. et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 54 , 1–12 (2022).
Waldegger, S. & Jentsch, T. Functional and structural analysis of CLC-K chloride channels involved in renal disease. J. Biol. Chem. 275 , 24527–33 (2000).
Matsumura, Y. et al. Overt nephrogenic diabetes insipidus in mice lacking the CLC-K1 chloride channel. Nat. Genet. 21 , 95–98 (1999).
Zhang, Q. et al. Exploring genes for immunoglobulin A nephropathy: a summary data-based mendelian randomization and fuma analysis. BMC Med. Genomics 16 , 16 (2023).
Wang, T. et al. Arachidonic acid metabolism and kidney inflammation. Int. J. Mol. Sci. 20 , 3683 (2019).
Das, U. Arachidonic acid in health and disease with focus on hypertension and diabetes mellitus. J. Adv. Res. 11 , 43–55 (2018).
Dent, C. I. et al. Quantifying splice-site usage: a simple yet powerful approach to analyze splicing. NAR Genomics Bioinforma. 3 , lqab041 (2021).
Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res. 50 , D988–D995 (2021).
Article PubMed Central Google Scholar
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) , 770–778 (IEEE Xplore, Las Vegas, 2016).
Li, G.-W. et al. Scapture: a deep learning-embedded pipeline that captures polyadenylation information from 3’ tag-based rna-seq of single cells. Genome Biol. 22 , 221 (2021).
Tay, Y., Dehghani, M., Bahri, D. & Metzler, D. Efficient transformers: a survey. ACM Comput. Surv . 55 , 1–28 (2022).
Tay, Y., Bahri, D., Yang, L., Metzler, D. & Juan, D.-C. Sparse sinkhorn attention. In International Conference on Machine Learning , 9438–9447 (PMLR, 2020).
Chennupati, S., Sistu, G., Yogamani, S. & A Rawashdeh, S. Multinet++: multi-stream feature aggregation and geometric loss strategy for multi-task learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops , 0–0 (IEEE Xplore, 2019).
Ling, J. P. et al. Ascot identifies key regulators of neuronal subtype-specific splicing. Nat. Commun. 11 , 137 (2020).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6 , 80–92 (2012).
Köhler, S. et al. The human phenotype ontology in 2021. Nucleic Acids Res. 49 , D1207–d1217 (2021).
Article ADS PubMed Google Scholar
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10 , 1523 (2019).
Article ADS PubMed PubMed Central Google Scholar
Dobin, A. et al. Star: ultrafast universal rna-seq aligner. Bioinformatics 29 , 15–21 (2013).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29 , 24–26 (2011).
Garrido-Martín, D., Palumbo, E., Guigó, R. & Breschi, A. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput Biol. 14 , e1006360 (2018).
Gargano, M. et al. The human phenotype ontology in 2024: phenotypes around the world. Nucleic Acids Res. 52 (2023).
You, N. et al. Splicetransformer predicts tissue-specific splicing linked to human diseases. Splicetransformer v1.0.0. https://doi.org/10.5281/zenodo.13824839 (2024).
Download references
Acknowledgements
We thank Prof. Yi Zhang, Prof. Haoxing Xu, and Prof. Haohong Li for helpful advice with manuscript writing. We thank Huijie Cao for helping with the model logo draft. We thank Yicheng Liu and Dr. Lina Liu for their helpful suggestions with manuscript preparation. We thank Xiaoyang Zhao and Asif Ashan for their assistance with gene network visualization. We thank you for the technical support by the Core Facilities of Liangzhu Laboratory. This work was supported by the Key R&D Program of Zhejiang (no. 2024SSYS0022), Special Funds of the National Natural Science Foundation of China (32141004), and the Open Project of Jiangsu Provincial Science and Technology Resources (Clinical Resources) Coordination Service Platform, no. TC2022B006. A subset of images (Figs. 1 a, 3 a, 4 a, and 6 a) were created with BioRender.com, with permission.
Author information
Authors and affiliations.
Department of Obstetrics and Gynecology of Sir Run Run Shaw Hospital & Liangzhu Laboratory, Zhejiang University School of Medicine, Hangzhou, China
Ningyuan You, Chang Liu, Hanying Jia, Tianyun Zhang, Shanshan Pei & Ning Shen
Institute of Genetics, Zhejiang University School of Medicine, Hangzhou, China
Yuxin Gu & Min-Xin Guan
Department of Hematology, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
National Clinical Research Center for Kidney Diseases, Jinling Hospital, Nanjing University School of Medicine, Nanjing, China
Song Jiang, Jinsong Shi & Zhihong Liu
Department of Bioinformatics, College of Life Sciences, Zhejiang University, Hangzhou, China
Research Institute of Intelligent Complex Systems, Fudan University, Shanghai, China
Bone Marrow Transplantation Center, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
Shanshan Pei
You can also search for this author in PubMed Google Scholar
Contributions
N.Y. designed and trained the model, collected data, performed data analysis, and drafted the original manuscript; C.L. drafted the original manuscript and revised the manuscript. Y.G. performed data analysis; R.W. performed data analysis and drafted the original manuscript; H.J. collected data, performed data analysis, and drafted the original manuscript; T.Z. performed data analysis; S.J. collected clinical samples and performed experiments to generate data regarding DN patients; J.S. performed data analysis; M.C., S.P., M.G., S.S., S.P. and Z.L. supervised the work and contributed to the manuscript; N.S. conceived, designed, and guided the study, and revised the manuscript with inputs from S.P., Z.L., and M.C. All authors read and approved the manuscript.
Corresponding authors
Correspondence to Zhihong Liu or Ning Shen .
Ethics declarations
Competing interests.
The authors have submitted a patent application for the method. Other than this, the authors declare that they do not have any competing interests.
Peer review
Peer review information.
Nature Communications thanks Dadi Gao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
You, N., Liu, C., Gu, Y. et al. SpliceTransformer predicts tissue-specific splicing linked to human diseases. Nat Commun 15 , 9129 (2024). https://doi.org/10.1038/s41467-024-53088-6
Download citation
Received : 27 November 2023
Accepted : 24 September 2024
Published : 23 October 2024
DOI : https://doi.org/10.1038/s41467-024-53088-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
About Nucleic Acids Research Editors & Editorial Board Information of Referees Alerts Self-Archiving Policy Dispatch Dates Advertising and Corporate Services ... Graphical Abstracts - Examples . Xrp1 governs the stress response program to spliceosome dysfunction .
Nucleic Acids Research is a peer reviewed fully open access journal publishing 24 issues per year online and in print. All papers published in the Journal are made freely available online under open access publishing agreements, with applicable charges. ... A video/graphical abstract is a visual representation of the central finding or ...
Nucleic Acids Research devotes its first issue each year to publishing information on online database resources of high value to the biological community. It contains: ... A graphical abstract is a visual representation of the central finding or methodology of the paper. It will be displayed in the table of contents and will be published as ...
Graphical abstract/eTOC; Unsolicited submissions considered; Commentary Commentaries should provide short descriptions and analyses of important research published in Molecular Therapy - Nucleic Acids. Include a declaration of interest statement before the references. Additional requirements include:
Template for Nucleic Acids Research (NAR) journal. Open as Template. View Source. View PDF. Author. Nucleic Acids Research. Last Updated. 5 years ago. License.
Graphical Abstract. Abstract. Nucleic acids are considered as fundamental molecules of living systems, which serve as universal genetic information messengers and repositories. ... Open Research. Data Availability Statement. Data sharing is not applicable to this article as no new data were created or analyzed in this study. References, ...
Nucleic Acids Res. 2016 Jul 27;44(13):e117. doi: 10.1093/nar/gkw430. Epub 2016 May 13. ... TSCAN has a graphical user interface (GUI) to support data visualization and user interaction. Furthermore, quantitative measures are developed to objectively evaluate and compare different pseudo-time reconstruction methods. ... Research Support, N.I.H ...
Nucleic Acids Res. 2023 Jul 21; 51(13): 6870-6882. Published online 2023 Jun 7. ... Graphical Abstract. Open in a separate window. Images in this article. Click on the image to see a larger version. ...
Nucleic Acids Res. 2023 Jun 23; 51(11): 5714-5742. Published online 2023 May 1. ... << Prev Graphical Abstract Next >> Graphical Abstract. Open in a separate window. RecF interacts with DnaN. Images in this article. Click on the image to see a larger version. ...
A graphical method is presented for displaying the patterns in a set of aligned sequences. The characters representing the sequence are stacked on top of each other for each position in the aligned sequences. The height of each letter is made proportional to its frequency, and the letters are sorted so the most common one is on top.
Nucleic Acids Research devotes a single issue in July to papers describing web-based software resources of value to the biological community. The Web Server Issue contains papers describing software programs that run on the web and provide useful computations on DNA, RNA and protein sequences or structures; analysis of high throughput ...
This method, called DRAGON (Determining Regulatory Associations using Graphical models on multi-Omic Networks), calibrates its parameters to achieve an optimal trade-off between the network's complexity and estimation accuracy, while explicitly accounting for the characteristics of each of the assessed omics 'layers.'.
Graphical abstractᅟ Electronic supplementary material The online version of this article (10.1007/s00216-018-1156-x) contains supplementary material, which is available to authorized users.
Graphical abstract. View. ... nucleic acids, lipids and carbohydrates in leukemia cells. Secondly, the same procedure was used to further detect different types of AML and found that Raman ...
Comparison of progr ams for 2D dra wing of nucleic acid structures. Solid dots indicate that a featur e is supported by a program, though the robustness of featur e implementations may vary betw ...
Abstract. The region surrounding a protein, known as the surface of interaction or molecular surface, can provide valuable insight into its function. Unfor
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a knowledge base for systematic analysis of gene functions in terms of the networks of genes and molecules. The major component of KEGG is the PATHWAY database that consists of graphical diagrams of biochemical pathways including most of the known me …
Zain and colleagues study clamp anti-gene oligonucleotides and improve their efficiency of invasion into double-strand DNA by introducing an intercalator. Additionally, they use these anti-gene clamps, targeting only a single 15-nt-long site in the huntingtin gene, to reduce the gene's expression in fibroblasts from a patient with Huntington disease.
YOU Research Group - Nucleic Acid Chemistry and Engineering. Department of Chemistry, University of Massachusetts, Amherst
Modular Fabrication of GFET Biosensors. In article number 2401796, Zhen Zhao, Jiahong Wang, Wenhua Zhou, and co-workers present a simple and robust graphene field-effect transistor biosensor fabricated through modular techniques, enabling a high-sensitive and specific detection of nucleic acids.This strategy also offers a possibility of adapting various microfluidic electronic devices with ...
INTRODUCTION. In nucleic acids research, particularly RNA research, structures are commonly drawn in 2-dimensional (2D) format ().Conventions such as the Leontis-Westhof notation are also important for denoting various types of canonical and non-canonical base-pairs and interactions in RNA structures.Since manually creating 2D structure drawings is laborious for large structures (), a variety ...
We present SpliceTransformer (SpTransformer), a deep-learning framework that predicts tissue-specific RNA splicing alterations linked to human diseases based on genomic sequence. SpTransformer ...