
- Why Does Water Expand When It Freezes
- Gold Foil Experiment
- Faraday Cage
- Oil Drop Experiment
- Magnetic Monopole
- Why Do Fireflies Light Up
- Types of Blood Cells With Their Structure, and Functions
- The Main Parts of a Plant With Their Functions
- Parts of a Flower With Their Structure and Functions
- Parts of a Leaf With Their Structure and Functions
- Why Does Ice Float on Water
- Why Does Oil Float on Water
- How Do Clouds Form
- What Causes Lightning
- How are Diamonds Made
- Types of Meteorites
- Types of Volcanoes
- Types of Rocks

The Endosymbiotic Theory
The endosymbiotic theory is a scientific theory that proposes that some of the organelles in the eukaryotic cells, such as mitochondria and chloroplast , have originated from free-living prokaryotes ( bacteria and archaea ). Endosymbiosis is the relationship between two organisms when one lives within the other organism, eventually benefiting both partners.
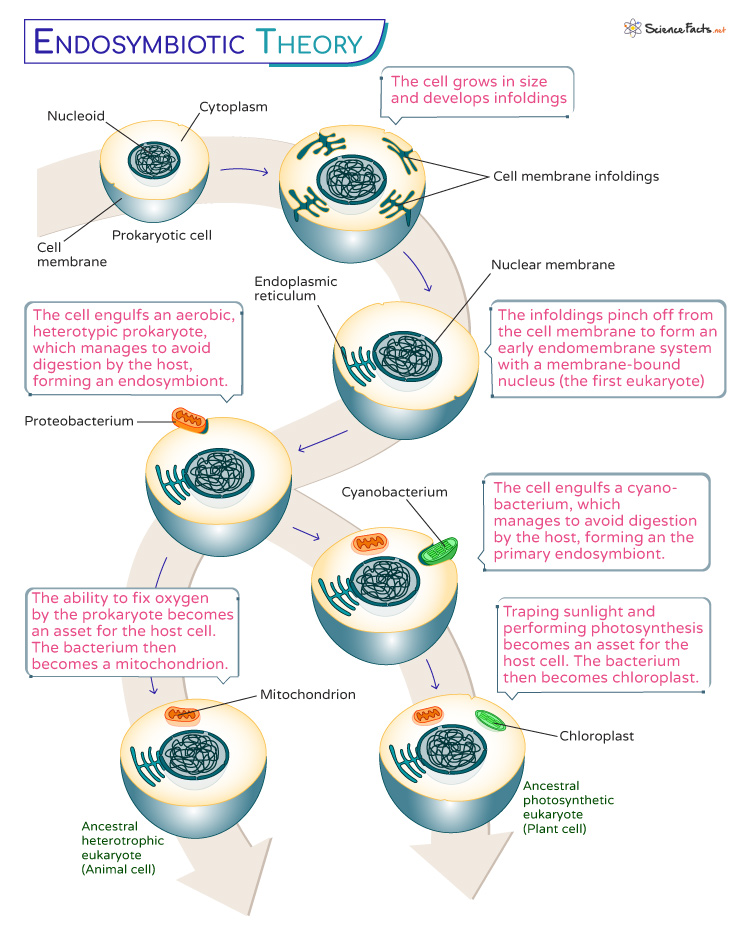
The endosymbiotic theory explains that when one organism, typically a microbe, takes up residence within the cell of another organism, over time, they form a close relationship that can be advantageous for both partners. The larger host cell provides a protected environment and essential nutrients. At the same time, the internalized microbe contributes its specialized functions, often becoming an organelle within the host cell.
The theory was conceptualized first by Konstantin Mereschkowsk in 1905 and then supported with evidence by Lynn Margulis in 1967.
The Theory of Endosymbiosis in Timeline
The German botanist Heinrich Anton de Bary coined the term ‘Symbiose’ to designate this coexistence. The concept of symbiosis , that two different organisms stably coexist and even give rise to a new type of organism, is attributed to Simon Schwendener.
19th Century
- In 1905, Russian biologist Konstantin Mereschkowski suggested that the origin of eukaryotic cells involved the engulfment of smaller prokaryotic cells.
- Around the same time, German botanist Andreas Schimper proposed that chloroplasts in plant cells might have originated from independent photosynthetic organisms.
1960s-1970s
- In 1967, Lynn Margulis proposed that eukaryotic cells, with their complex structures and organelles, resulted from symbiotic relationships between ancestral prokaryotic cells. He argued that mitochondria, the cellular powerhouses, were once free-living bacteria capable of aerobic respiration . Similarly, she suggested that chloroplasts originated from photosynthetic bacteria that became incorporated into host cells.
1980s-1990s
- Researchers discovered striking similarities between the DNA of organelles like mitochondria and chloroplasts and the DNA of modern-day bacteria.
- Protists like single-celled organisms are found to host certain bacteria fixing nitrogen (nitrogen-fixing bacteria).
2000s-Present
- In the 21st Century, endosymbiosis forms the basis of the origin of mitochondria, chloroplast, and other organelles. It also explains the development of complex multicellular organisms and the coevolution of symbiotic partners.
The Process of Endosymbiosis
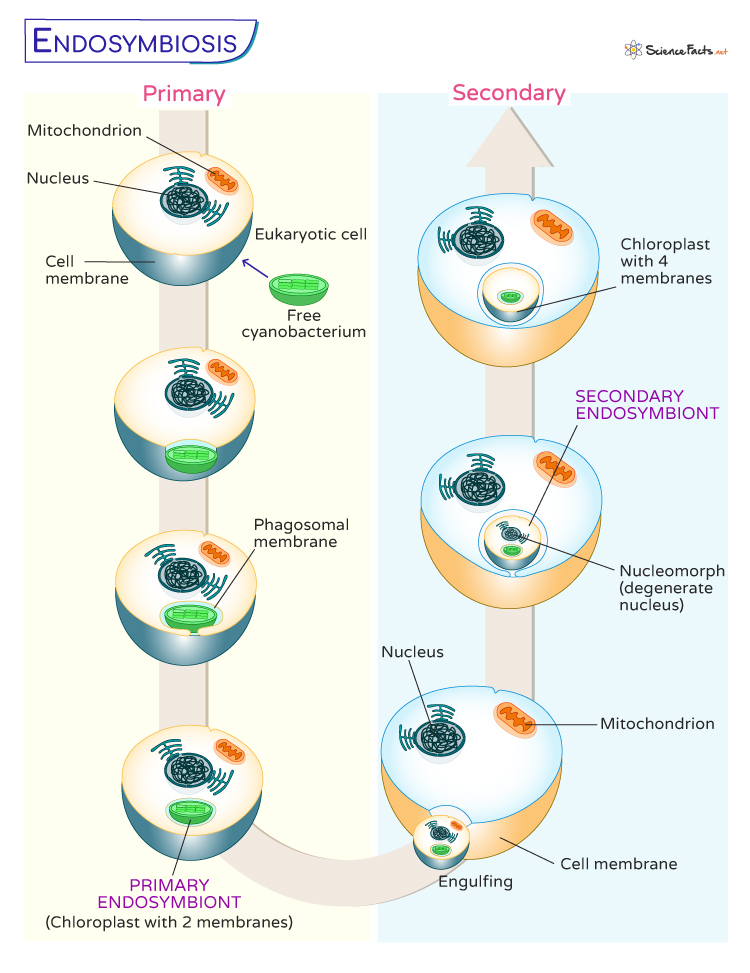
According to the endosymbiotic theory, the symbiotic origin of eukaryotic cells is a multi-event process.
1. Primary Endosymbiosis
It refers to the initial engulfment of a free-living bacterium by a host cell, creating a new organelle within the host cell. The most prominent examples of primary endosymbiosis are the origin of mitochondria and chloroplasts; both were once free-living cells. Here, two membranes surround the organelles; the inner is obtained from the bacterium, and the outer is derived from the host cell.
- Origin of Mitochondria : A eukaryotic cell engulfed a bacterium capable of aerobic respiration. This bacterium provides a valuable energy source through respiration. Over time, the host cell and the engulfed bacterium developed a mutually beneficial relationship. The bacterium became the mitochondrion, retaining its own DNA and membrane structure while working in tandem with the host cell.
- Origin of Chloroplasts : Here, an ancestral host cell captured a photosynthetic bacterium, which could convert sunlight into energy through photosynthesis . As with mitochondria, a symbiotic partnership formed, with the photosynthetic bacterium evolving into the chloroplast. It allowed the host cell to harness the power of photosynthesis for energy production.
2. Secondary Endosymbiosis
It involves a eukaryotic cell engulfing another eukaryotic cell already undergoing primary endosymbiosis. This secondary engulfment results in a more complex cellular arrangement, leading to the diversification of eukaryotic lineages and the emergence of new types of organelles.
- Formation of Plastids : These organelles involved in photosynthesis are found in various algae and plants. Different groups of algae have acquired plastids through secondary endosymbiosis, which consists of the engulfment of photosynthetic eukaryotic cells. In contrast to the two membranes of primary organelles, four membranes surround chloroplasts obtained by secondary endosymbiosis. In most cases, the nucleus of the engulfed cell disappears, with the remains of this nucleus still found lying between the two pairs of membranes. This structure is called a nucleomorph.
Thus, the endosymbiotic theory explains the presence of double-membraned organelles within protists.
Evidence that Supports the Endosymbiotic Theory
There are several proofs to support the Endosymbiotic Theory. However, the discovery of independent DNA (from the host) in mitochondria and chloroplasts supported the theory the most. The other evidences are as follows:
- Structural Similarities : Mitochondria and chloroplasts share structural characteristics with free-living bacteria, such as double membranes and DNA. Both the organelles are almost of the same size as the bacterial cell.
- Reproduction : Mitochondria and chloroplasts replicate within the cell independently, similar to how bacteria reproduce.
- Genetic Evidence : The DNA within mitochondria and chloroplasts is more similar to bacterial DNA than the host cell’s nucleus.
- Evolutionary Relationships : Analysis of genetic sequences shows that mitochondria and chloroplasts are more closely related to specific groups of bacteria than eukaryotic cells.
However, the statement that mitochondria and chloroplasts are much larger than prokaryotic cells does not support the endosymbiotic theory.
What is the Importance of the Endosymbiotic Theory
It is important because the theory explains the origin of the eukaryotic cells. It also describes how chloroplast and mitochondria might have originated from once free-living prokaryotes. This understanding has reshaped our perception of how fundamental cellular organelles came to be and how multicellular life forms arose.
The endosymbiotic theory also highlights how cooperation and symbiosis have played pivotal roles in shaping cellular evolution.
- Endosymbiosis – Ib.bioninja.com.au
- Endosymbiosis and the Evolution of Eukaryotes – Bio.libretexts.org
- Endosymbiotic Theories forEukaryote Origin – Royalsocietypublishing.org
- Endosymbiosis – Evolution.berkeley.edu
- Endosymbiosis: The Feeling is Not Mutual – Ncbi.nlm.nih.gov
- Endosymbiosis – Cell.com
Article was last reviewed on Tuesday, October 3, 2023
Related articles
Leave a reply cancel reply.
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
Popular Articles
Join our Newsletter
Fill your E-mail Address
Related Worksheets
- Privacy Policy
© 2024 ( Science Facts ). All rights reserved. Reproduction in whole or in part without permission is prohibited.

Science Hypothesis
Ai generator.
Hypothesis are the bedrock of scientific investigation, guiding researchers toward understanding the unknown. Crafting effective science hypotheses involves precise formulation and prediction. This hypothesis statement guide delves into the intricacies of constructing science hypothesis statements, offering practical examples and valuable tips to ensure your hypothesis stand strong against the rigors of experimentation and analysis.

What is Science Hypothesis? – Definition
A science hypothesis is a proposed explanation or educated guess that can be tested through experimentation or observation. It serves as a preliminary assumption or prediction about a phenomenon, often derived from existing knowledge or theories. Science hypotheses are essential for guiding research and helping scientists investigate the validity of their predictions.
What is an example of a hypothesis statement in science?
Example of a hypothesis statement in science: “If the temperature of water increases, then the rate of plant growth will also increase.” This hypothesis predicts a cause-and-effect relationship between water temperature and plant growth, which can be tested through controlled experiments.
100 Science Hypothesis Statement Examples

Size: 223 KB
Science hypotheses lay the foundation for empirical exploration. These Thesis statements predict outcomes based on existing knowledge and guide research. Explore a variety of science hypothesis examples across different disciplines, showcasing the diverse ways scientists propose, test, and validate their assumptions. From physics to biology, chemistry to astronomy, delve into these examples that highlight the essence of scientific inquiry and discovery.
- Physics : If the mass of an object increases, its gravitational pull on another object will also increase.
- Biology : If plants are exposed to different light wavelengths, then the one exposed to red light will exhibit the highest growth rate.
- Chemistry : If the concentration of a reactant increases, then the rate of the chemical reaction will also increase.
- Astronomy : If the distance between two galaxies decreases, then their gravitational attraction will intensify.
- Geology : If the temperature of a rock sample increases, then its density will decrease due to expansion.
- Psychology : If individuals are exposed to positive affirmations, then their self-esteem scores will improve.
- Sociology : If economic inequality increases, then crime rates within a community will also rise.
- Environmental Science : If pollution levels decrease in a river, then the diversity of aquatic species will increase.
- Computer Science : If the processing speed of a computer chip increases, then the execution time of a software program will decrease.
- Meteorology : If atmospheric pressure drops significantly, then the likelihood of stormy weather conditions will rise.
- Neuroscience : If individuals engage in regular meditation, then their brain’s gray matter volume in regions associated with mindfulness will increase.
- Economics : If interest rates decrease, then consumer spending will rise due to increased borrowing.
- Anthropology : If a society’s cultural diversity increases, then its acceptance of differing norms and values will also grow.
- Zoology : If predators are introduced to an ecosystem, then the population of prey species will decline.
- Medical Research : If a new drug is administered, then patients with a specific medical condition will experience a reduction in symptoms.
- Nutrition Science : If individuals consume a diet high in antioxidants, then their risk of developing certain chronic diseases will decrease.
- Materials Science : If the temperature of a metal is lowered, then its electrical conductivity will decrease due to reduced kinetic energy.
- Political Science : If voter education initiatives increase, then voter turnout rates in elections will also rise.
- Geography : If urbanization expands in a region, then the average local temperature will increase due to the heat island effect.
- Ecology : If a keystone species is removed from an ecosystem, then the overall biodiversity of that ecosystem will be negatively impacted.
- Medieval History : If trade routes between two civilizations strengthen, then cultural exchange and technological advancements will flourish.
- Microbiology : If a specific bacterium is introduced to a microbial community, then it will outcompete other species for resources.
- Oceanography : If ocean temperatures rise, then coral reefs will experience bleaching due to the loss of symbiotic algae.
- Education : If class sizes are reduced, then student engagement and learning outcomes will improve.
- Genetics : If individuals inherit two recessive alleles for a particular trait, then they will exhibit the trait phenotypically.
- Criminology : If community policing initiatives are implemented, then the crime rate in neighborhoods will decrease due to improved trust between law enforcement and residents.
- Botany : If plants are exposed to varying levels of nutrients, then their growth rate and overall health will be affected accordingly.
- Epidemiology : If individuals are vaccinated against a specific virus, then the incidence of that virus in the population will decline.
- Architecture : If buildings are designed with energy-efficient features, then their energy consumption and environmental impact will be reduced.
- Literary Studies : If readers are exposed to diverse genres of literature, then their vocabulary and literary comprehension will expand.
- Mechanical Engineering : If the surface area of a heat exchanger is increased, then its efficiency in transferring thermal energy will improve.
- Artificial Intelligence : If a machine learning algorithm is trained on a larger dataset, then its accuracy in making predictions will increase.
- Sports Science : If athletes incorporate specific pre-game rituals, then their performance and focus during competitions will improve.
- Archaeology : If a new excavation site is discovered, then artifacts and evidence of past civilizations will be uncovered.
- Film Studies : If films use non-linear storytelling techniques, then audience engagement and interpretation will become more complex.
- Fashion Design : If clothing materials with better breathability are used, then wearers’ comfort levels in hot weather will increase.
- Music Psychology : If listeners are exposed to music with a fast tempo, then their heart rate and energy levels will be positively affected.
- Environmental Engineering : If a wastewater treatment system is upgraded, then the water quality of nearby rivers and streams will improve.
- Philosophy : If ethical dilemmas are discussed openly, then individuals’ moral reasoning and decision-making skills will become more refined.
- Cognitive Science : If individuals practice mindfulness meditation, then their attention span and cognitive control will enhance.
- Political Economy : If trade barriers between two countries are lifted, then their economic interdependence and cooperation will strengthen.
- Agricultural Science : If certain crops are rotated in a field, then soil fertility and nutrient content will be better maintained.
- Cultural Anthropology : If cultural norms change to value gender equality, then the division of labor and social roles will evolve accordingly.
- Linguistics : If a language’s phonetic structure is altered, then the perception and articulation of speech sounds will be affected.
- Religious Studies : If religious festivals are celebrated widely, then social cohesion and a sense of community among participants will increase.
- Urban Planning : If public transportation infrastructure is improved, then the use of private vehicles and traffic congestion will decrease.
- Renewable Energy : If solar panel efficiency increases, then the cost-effectiveness of solar energy as a power source will improve.
- Sustainable Agriculture : If organic farming practices are adopted, then soil health and biodiversity in agricultural fields will be enhanced.
- Human Genetics : If a specific gene mutation is present, then the likelihood of developing a hereditary disease will be higher.
- Space Exploration : If a spacecraft is sent to a distant planet, then the data collected will provide insights into its composition and environment.
- Cultural Studies : If a society values inclusivity in its media representations, then stereotypes and biases will be challenged.
- Quantum Physics : If two entangled particles are measured, then the measurement of one particle will instantaneously affect the state of the other particle, regardless of distance.
- Social Work : If support systems are established for individuals facing addiction, then their likelihood of successful recovery will increase.
- Civil Engineering : If a bridge is constructed using specific materials and design principles, then its load-bearing capacity and structural integrity will be maximized.
- Educational Technology : If interactive learning platforms are integrated into classrooms, then students’ engagement and retention of concepts will rise.
- Animal Behavior : If a specific stimulus is introduced to an animal’s environment, then its behavioral response will indicate whether the stimulus is perceived as positive or negative.
- Public Health : If a vaccination campaign targets a high percentage of the population, then the spread of a contagious disease will be curbed.
- Forensic Science : If DNA evidence is analyzed from a crime scene, then it can be matched to potential suspects or used to exonerate individuals.
- Game Design : If a game incorporates branching storylines, then players’ choices will lead to multiple possible outcomes and endings.
- Gender Studies : If gender stereotypes are challenged in educational settings, then students’ understanding of gender roles and identities will evolve.
- Particle Physics : If a new particle is discovered in particle accelerator experiments, then it may contribute to our understanding of fundamental forces.
- Culinary Science : If cooking techniques are adjusted, then the texture and flavor of a dish will be enhanced.
- Developmental Psychology : If children are exposed to early childhood education programs, then their cognitive and social development will be positively influenced.
- Journalism : If journalists provide unbiased coverage of events, then the public’s perception and understanding of news stories will be more accurate.
- Business Management : If a company implements remote work policies, then employees’ job satisfaction and productivity will be impacted.
- Astronomy : If a telescope observes a distant celestial object, then its light spectrum can reveal information about its composition and distance.
- Climate Science : If greenhouse gas emissions continue to rise, then global temperatures will increase, leading to more frequent and severe climate events.
- Molecular Biology : If a specific gene is mutated, then the protein it codes for may lose its function, leading to a genetic disorder.
- Urban Sociology : If urban planning focuses on mixed-use development, then neighborhoods will become more walkable and vibrant.
- Environmental Science : If deforestation continues in a particular region, then biodiversity loss and habitat destruction will result.
- Educational Psychology : If students receive constructive feedback, then their academic performance and self-esteem will improve.
- Sports Nutrition : If athletes consume a balanced diet, then their energy levels and physical performance will be optimized.
- Industrial Engineering : If a manufacturing process is streamlined, then production efficiency and cost-effectiveness will increase.
- Climate Change Mitigation : If renewable energy sources replace fossil fuels, then carbon emissions and air pollution will decrease.
- Criminal Justice : If restorative justice programs are implemented, then recidivism rates among offenders will decrease.
- Cognitive Neuroscience : If brain imaging techniques are used, then neural activity patterns associated with memory retrieval can be identified.
- Environmental Policy : If conservation policies are enforced, then endangered species populations will have a chance to recover.
- Tourism Management : If sustainable tourism practices are adopted, then the negative impact of tourism on local ecosystems will be minimized.
- Public Opinion Research : If surveys are conducted on political preferences, then insights into voter behavior and attitudes can be gained.
- Sociolinguistics : If language use changes over time, then linguistic patterns and dialects in a community may evolve.
- Consumer Behavior : If marketing strategies incorporate social media influencers, then consumer purchasing decisions will be influenced.
- Digital Communication : If online privacy measures are strengthened, then users’ data security and trust in digital platforms will increase.
- Cancer Research : If a specific genetic mutation is identified, then targeted therapies can be developed to treat the cancer associated with that mutation.
- Human Rights Advocacy : If educational campaigns raise awareness about human rights violations, then public pressure on governments to address these issues will rise.
- Educational Assessment : If standardized tests are redesigned to focus on critical thinking skills, then students’ analytical abilities will be better evaluated.
- Epidemiology : If a specific virus spreads within a community, then the rate of infection and transmission can be studied to develop effective containment strategies.
- Cognitive Psychology : If memory recall is examined under different conditions, then the factors influencing memory retrieval can be identified.
- Financial Economics : If interest rates are lowered by the central bank, then borrowing costs for businesses and individuals will decrease.
- Marine Biology : If ocean temperatures rise due to climate change, then coral bleaching events will become more frequent, leading to coral reef degradation.
- Political Science : If voter turnout is influenced by campaign advertising, then the correlation between media exposure and voting behavior can be analyzed.
- Clinical Psychology : If cognitive-behavioral therapy is administered to individuals with anxiety disorders, then their symptoms will show a reduction.
- Public Policy : If a government enforces stricter regulations on smoking in public spaces, then the prevalence of smoking-related health issues will decline.
- Material Science : If a new material is developed with specific properties, then its potential applications in various industries can be explored.
- Language Acquisition : If children are exposed to multiple languages in their early years, then their linguistic skills may develop differently compared to monolingual children.
- Tourism Economics : If travel restrictions are lifted, then the recovery of the tourism industry and its contribution to the local economy can be assessed.
- Behavioral Economics : If individuals are given incentives to make environmentally friendly choices, then the impact of economic incentives on behavior can be studied.
- Educational Technology : If online learning platforms are used in classrooms, then their effect on student engagement and academic performance can be evaluated.
- Health Policy : If universal healthcare coverage is implemented, then access to medical services and health outcomes for the population can be improved.
- Agricultural Economics : If crop yields are compared between traditional farming methods and modern agricultural practices, then the efficiency of different approaches can be determined.
- Literary Analysis : If a specific theme is analyzed across different literary works, then the ways in which authors address and convey that theme can be explored.
Science Hypothesis Statement Examples for Psychology
These psychology hypothesis pertain to human behaviors, emotions, or cognitive processes. They are tailored to the field of psychology, which studies the human mind and behavior. For instance, “Effects of Sleep on Memory” posits a connection between sleep duration and memory performance.
- Effects of Sleep on Memory : People who sleep 8 hours per night will perform better on memory tests compared to those who sleep only 4 hours.
- Role of Colors in Mood Regulation : Exposure to blue light will decrease feelings of sadness in depressed individuals.
- Childhood Attachment and Adult Relationships : Individuals with secure childhood attachments will have more stable romantic relationships in adulthood.
- Influence of Music on Productivity : Listening to classical music while working increases task completion rates among office workers.
- Gaming and Reaction Time : Regular gamers will have quicker reaction times than non-gamers in response to unexpected stimuli.
- Effects of Meditation on Stress : Individuals who practice daily meditation will report lower stress levels compared to those who don’t meditate.
- Social Media Usage and Loneliness : High usage of social media correlates with increased feelings of loneliness in teenagers.
- Class Size and Student Performance : Students in smaller class sizes will score higher on standardized tests than students in larger class sizes.
- Scent and Memory Recall : People exposed to a specific scent during learning will recall information better when the same scent is present during retrieval.
- Financial Incentives and Motivation : Providing financial incentives will increase motivation for completing mundane tasks.
Simple Science Hypothesis Statement Examples
These are basic and straightforward scientific hypotheses that cover various fields, such as biology or physics. They’re easy to understand even for people without much scientific background. For instance, the simple hypothesis tatement about “Plant Growth” directly relates the use of fertilizer to plant height.
- Plant Growth : Adding fertilizer will make plants grow taller.
- Solar Energy : Increasing sunlight exposure will increase the voltage output of a solar cell.
- Density : Objects made of metal will sink in water.
- Digestion : Enzyme supplements will increase the speed of food digestion.
- Osmosis : Potatoes placed in salt water will shrink due to loss of water.
- Evaporation : Water will evaporate faster on a hot day compared to a cold day.
- Nutrition : Plants given sugar water will develop yellow leaves.
- Magnetism : Increasing the temperature of a magnet will decrease its magnetic strength.
- Conduction : Metals will conduct electricity better than plastics.
- Reflection : Shiny surfaces reflect more light than dull surfaces.
Strong Science Hypothesis Statement Examples
These are more detailed and specific hypotheses, often relating to a well-defined scientific question. They may also suggest a precise outcome or relationship. For example, “Vaccination and Immunity” indicates a specific result (production of specific antibodies) in response to a defined action (vaccinating mice).
- Environmental Toxins and Cell Growth : Exposure to specific environmental toxins will inhibit the division of cells in an organism.
- Nutrition and Cognitive Performance : Diets rich in omega-3 fatty acids will significantly enhance cognitive performance in adults over 60.
- Genetic Mutations and Disease Resistance : Specific genetic mutations in fruit flies will confer resistance to a particular pesticide.
- Neurotransmitters and Behavior : An increase in serotonin levels in the brain will lead to a decrease in aggressive behaviors in rats.
- Plant Pathogens and Resistance : Tomato plants genetically modified to express the XYZ gene will resist infection from the ABC pathogen more effectively than non-modified plants.
- Vaccination and Immunity : Vaccinating mice with a particular strain of virus will lead to the production of specific antibodies that prevent future infections.
- Hormonal Levels and Bone Density : Post-menopausal women with decreased estrogen levels will have a significant reduction in bone density compared to pre-menopausal women.
- Enzyme Concentration and Reaction Rate : Doubling the concentration of an enzyme in a solution will double the rate of the substrate’s conversion to the product.
- Climate Change and Coral Bleaching : An increase in sea surface temperature by 2°C will lead to a 50% increase in coral bleaching events.
- Pesticides and Pollinator Health : Exposure to the pesticide DEF will reduce the foraging ability of honeybees by at least 30%.
Scientific Hypothesis Statement Examples
These are broader scientific hypothesis applicable to different scientific disciplines. They’re structured to make clear, testable predictions about the relationship between variables. “Bacterial Growth,” for instance, predicts the outcome of bacteria exposed to UV light.
- Bacterial Growth : Bacteria exposed to ultraviolet (UV) light will have a reduced growth rate compared to those not exposed to UV light.
- Antibiotic Resistance : Overuse of antibiotics in livestock will lead to an increase in antibiotic-resistant bacteria in humans.
- Evolutionary Adaptation : Birds with longer beaks will have an advantage in accessing food after a drastic environmental change.
- Photosynthesis Rate : Plants grown under red light will have a lower rate of photosynthesis compared to those grown under blue light.
- Stem Cell Differentiation : The presence of growth factor X will guide stem cells to differentiate into nerve cells more frequently than muscle cells.
- Ozone Layer and UV Radiation : Depletion of the ozone layer will result in increased UV radiation levels on Earth’s surface.
- Protein Folding : Mutation at position 123 in protein Z will lead to a misfolded protein structure.
- Water Quality and Fish Health : Rivers with high levels of industrial pollutants will have a reduced fish population due to compromised gill functionality.
- Seismic Activity and Plate Tectonics : Regions located at the boundaries of tectonic plates will experience more frequent and stronger earthquakes.
- Drug Efficacy : Patients treated with drug Y will recover from infection twice as fast as those treated with a placebo.
Alternative Hypothesis Statement Examples for Science
The alternative hypothesis states that there is a statistically significant relationship between two variables. It’s what you might want to prove or demonstrate. For example, the hypothesis about “Green Tea and Metabolism” suggests that drinking green tea can have a positive effect on metabolic rates.
- Dietary Supplements and Energy Levels : Consuming a daily vitamin B12 supplement will increase energy levels in vegans.
- Soil Type and Crop Yield : Sandy soil will produce a lower maize yield than loamy soil.
- Air Pollution and Respiratory Diseases : Living in areas with higher particulate matter (PM2.5) levels will increase the incidence of respiratory diseases.
- Green Tea and Metabolism : Drinking green tea daily will increase metabolic rates in adults.
- Exercise and Brain Health : Engaging in regular aerobic exercise will increase cognitive function in older adults.
- Artificial Sweeteners and Appetite : Consuming artificial sweeteners will increase appetite in individuals.
- Forest Density and Wildlife Diversity : Forests with higher tree density will support a more diverse range of wildlife.
- Hydration and Skin Health : Drinking at least 2 liters of water daily will improve skin elasticity.
- Biofuels and Engine Performance : Engines running on biofuel will have a higher fuel efficiency than those running on traditional petroleum fuels.
- Artificial Light and Plant Growth : Plants grown under LED lights will have a faster growth rate than those grown under fluorescent lights.
Null Hypothesis Statement Examples for Science
The null hypothesis posits that there is no relationship between two variables. It’s the statement you want to test against. Scientists often set out to reject the null hypothesis to demonstrate there’s a relationship. For instance, “Diet and Weight Loss” asserts there’s no difference in weight loss outcomes between two diet types.
- Diet and Weight Loss : There is no difference in weight loss between individuals on a low-carb diet and those on a low-fat diet.
- Antibacterial Soap and Hand Hygiene : Using antibacterial soap does not decrease the number of bacteria on hands compared to using regular soap.
- Meditation and Blood Pressure : There is no difference in blood pressure levels between individuals who meditate daily and those who don’t.
- Organic Foods and Nutrient Content : Organic fruits and vegetables have the same nutrient content as non-organic fruits and vegetables.
- Pain Relievers and Pain Reduction : Over-the-counter pain reliever X does not reduce pain more effectively than a placebo.
- Educational Method and Learning : There is no difference in learning outcomes between students taught using method A and those taught using method B.
- Herbal Treatment and Sleep Duration : Herbal treatment Y does not increase sleep duration compared to a placebo.
- Sunscreen and Sunburn : There is no difference in sunburn incidence between individuals using sunscreen with SPF 30 and those using sunscreen with SPF 50.
- Caffeine and Alertness : Consuming caffeine does not increase alertness levels compared to not consuming caffeine.
- Probiotics and Gut Health : Taking daily probiotics does not increase the diversity of gut bacteria compared to not taking probiotics.
What is a good hypothesis for a science project?
A good hypothesis is a fundamental cornerstone for any scientific project. It provides direction for your research, helping you to focus your investigations and understand the potential outcomes. Here’s what characterizes a good hypothesis:
- Testable : A good hypothesis must be something that can be supported or refuted through experimentation, observation, or analysis.
- Clear and Concise : It should be straightforward and to the point, making it easier for you or others to test.
- Logical : It should make logical sense, building upon existing knowledge and literature.
- Specific : The hypothesis should clearly identify the variables and the relationship between them.
- Relevant : It should be pertinent to the subject matter and not diverge into unrelated areas.
- Predictive : It should make a clear prediction about what you expect to happen in your study.
How do you write a scientific hypothesis statement? – A Step by Step Guide
- Identify Your Research Question : Before you can draft a hypothesis, you need to determine what you’re trying to answer. For example, “Does the type of soil affect plant growth?”
- Perform Preliminary Research : Understand existing literature on the topic. This will help ensure that your hypothesis is original and rooted in current understanding.
- Independent Variable (what you change): e.g., type of soil.
- Dependent Variable (what you measure): e.g., plant growth.
- Make a Prediction : Based on your research, predict the relationship between your variables.
- If : Describes the change or treatment (independent variable).
- Then : Predicts the outcome (dependent variable).
- Because : Provides a rationale based on your background research. E.g., “If a plant is grown in sandy soil, then it will grow slower than in loamy soil, because sandy soil retains less water.”
- Keep it Simple : Avoid complex sentences or jargon. Your hypothesis should be understandable even to someone not in your field.
- Review and Revise : Once drafted, revisit your hypothesis. Ensure it aligns with your research question and that it remains clear and testable.
Tips for Writing Science Hypothesis
- Start with Curiosity : Your initial question should stem from genuine curiosity. It might begin as a broad query which you then refine.
- Use Open-Ended Questions : Start your question with words like “How,” “What,” or “Why.” These types of questions don’t presuppose an answer and lead to more in-depth investigation.
- One Variable at a Time : Especially for beginner projects, limit your hypothesis to one independent variable to keep your study focused and manageable.
- Avoid Biased Language : Your hypothesis should not show any personal biases. Instead of “I believe” or “I think,” use neutral terms.
- Stay Relevant to Available Tools and Resources : Ensure that you can test your hypothesis with the tools, time, and resources available to you.
- Peer Review : Before finalizing your question and hypothesis, have a peer or mentor review it. They might catch ambiguities or complexities you missed.
- Be Ready to Accept Any Outcome : A common mistake is becoming too attached to proving your hypothesis right. Remember, disproving a hypothesis can be just as valuable as proving it.
By carefully crafting your research question and hypothesis, you’ll set a solid foundation for your science project. Whether your results support or challenge your initial predictions, you’ll contribute to the vast and ever-growing body of scientific knowledge.
Text prompt
- Instructive
- Professional
10 Examples of Public speaking
20 Examples of Gas lighting

IMAGES
VIDEO
COMMENTS
In other words, the capacity of bacteria to adapt is such that if it is to their advantage to influence their host preferences for food, they will. Here we explore the hypothesis that there is a positive-feedback relationship between the composition of the gut microbiota and food preferences.
A hypothesis in biology is a critical component of scientific research that proposes an explanation for a specific biological phenomenon. Writing a well-formulated hypothesis sets the foundation for conducting …
scientific hypothesis, an idea that proposes a tentative explanation about a phenomenon or a narrow set of phenomena observed in the natural world. The two primary features of a scientific hypothesis are …
A hypothesis is a tentative, testable answer to a scientific question. Once a scientist has a scientific question she is interested in, the scientist reads up to find out what is already known …
Learn how to write a strong hypothesis with our comprehensive guide. Step-by-step techniques with examples to formulate clear, testable hypotheses that lay the foundation for successful research. Ideal for students, academics, and …
A science hypothesis is a proposed explanation or educated guess that can be tested through experimentation or observation. It serves as a preliminary assumption or prediction about a phenomenon, often derived from …
Hypothesis is an educated guess or proposed explanation for a phenomenon, based on some initial observations or data. It is a tentative statement that can be tested and …